For many decades, the only way to detect sepsis – bacterial growth in blood – was isolating the bacteria and growing bacterial colonies on a special medium. This was done by first spinning down the blood, which brought the blood cells and bacteria into the pellet. The pellet was spread on a blood agar plate and the researcher had to wait for bacterial colonies to grow. This older method is limited by time commitment (takes between 1-7 days to complete), and the fact that many bacteria cannot be cultured on standard media1.
Modern methods of bacterial detection in blood include isolation and detection of bacterial nucleic acids. Indeed, most bacteria are detected and classified by their 16S rRNA, but the same upstream methods could be used to isolate bacterial DNA. In this article, I am going to introduce you to three main techniques to isolate bacterial nucleic acids from blood: (1) direct isolation; (2) centrifugation and filtration; and (3) affinity pull-down.
1. Direct Isolation
The main problem of isolating RNA from human blood is a high concentration of RNAses present in blood cells. If you try to lyse blood cells and then isolate bacterial RNA, you will get a low yield because RNAses released from blood cells will degrade all RNA types. You need to perform the lysis in conditions that will inactivate RNAses; for example, using RNAprotect Cell Reagent from Qiagen2. After the lysis, you can reverse transcribe RNA using specific primers.
2. Centrifugation and Filtration
Most of the modern methods of bacterial nucleic acid from blood are based on the separation of blood and bacterial cells. This prevents the signal from bacterial RNA from being masked by the mass of human RNA.
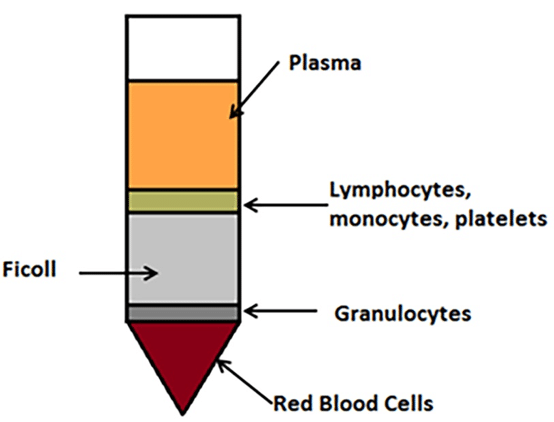
Red blood cells have a diameter of 8 -15 ?m, while bacterial cells are around 1 ?m. A number of separation protocols rely on Ficoll-Paque buffer (Sigma-Aldrich). The blood is either layered on top of the buffer or mixed with the buffer and centrifuged on low speed ~380 xg. The red blood cells that constitute the main part of the blood cells sediment while bacteria remain suspended in the buffer (Figure 1). The entire gradient is then filtered through a 0.22 ?m membrane that doesn’t allow bacteria cells to pass through. Collected bacteria can be used to isolate DNA or RNA3.
3. Affinity Pull-Down
When you experience an infection, one immunological response is the production of antibodies that are specific for an antigen expressed on the surface of the infiltrating pathogen. Biologists have exploited this antibody:antigen interaction in a number of laboratory techniques, including affinity pull-downs for bacterial cells.
In an affinity pull-down, magnetic or sepharose beads covered in antibodies or binding molecules that specifically bind surface antigens of bacteria are added to a bacteria-containing blood sample. The strength of the antibody/molecule:antigen bond interactions (aka affinity) keeps the magnetic bead and bacteria “stuck” together. The magnet is then applied to a sample. The beads are pulled down by the magnetic field, and the sample is washed to dispose of any contaminants. The magnetic bead: bacteria complexes treated with high salt buffer to break the bonds, and are further separated by another magnet application, releasing the bacteria cells while keeping the magnetic beads in place.
An example of a binding molecule that is often used in affinity pull-downs is mannose-binding lectin. This molecule binds to the surface of all clinical pathogens of interest in antibiotic resistance situations, including enterobacteria and streptococci, as well as many other bacteria and viruses4.
Alternatively, you can magnetically capture the human cells, leaving the bacteria to be filtered out from plasma. Magnetic beads coated with anti-human CD45 antibody reduced the number of human cells by 98%5.
In conclusion, there is a number of methods to separate bacteria from blood. Which one to choose depends on your laboratory setup and your preference.
Sources:
- Pitt W.G. et al. (2016) Rapid separation of bacteria from blood-review and outlook. Biotechnol Prog. 8;32(4):823-39. doi: 10.1002/btpr.2299.
- Brennecke J. et al. (2017) High-yield extraction of Escherichia coli RNA from human whole blood. J Med Microbiol. 66(3):301-311. doi: 1099/jmm.0.000439.
- D’Amico L. et al. (2017) Isolation and concentration of bacteria from blood using microfluidic membraneless dialysis and dielectrophoresis Lab Chip. 17(7): 1340–1348. doi: 1039/c6lc01277a
- Kang J.H. et al. (2014) An extracorporeal blood?cleansing device for sepsis therapy. Nat Med; 20:1211–1216.
- Vutukuru M.R. et al. (2016) A rapid, highly sensitive and culture-free detection of pathogens from whole blood by removal of white blood cells using immuno-magnetic beads. J Microbiol Methods.127:59-61. doi: 1016/j.mimet.2016.05.018.