For most molecular biology purposes, DNA is thought of as a string of nucleotides running from 3’ to 5’, and the corresponding mRNA sequence is complementary to this DNA string. However, visualizing this quirky DNA structure for what it is – two antiparallel strands joined together – it quite important for many applications, such as whole transcriptome analysis.
The Nomenclature
So, let’s recap. After the DNA double helix is separated into two strands by the RNA polymerase, one of the strands serves as RNA template. Depending on the transcription promoter (located on only one DNA strand), the majority of mRNA is transcribed using an antisense/Watson/template DNA strand that reads from 3’ to 5’. The molecule of RNA that protein is read from is called sense or “+” strand, whereas the opposite strand is antisense or “-”. For example, consider the DNA fragment:
3′ CGCTATAGCGTTT 5′ + strand
5′ GCGATATCGCAAA 3′ – strand
| Example sequence | Description | Function |
|---|---|---|
| 3′–CGCTATAGCGTTT–5′ | DNA antisense strand or Watson strand (template/non-coding) | Template for transcription. |
| 5′–GCGATATCGCAAA–3′ | DNA sense strand or Crick strand (non-template/coding) | Complementary to the template strand. |
| 5′–GCGAUAUCGCAAA–3′ | mRNA sense transcript | RNA strand transcribed from the Watson (template/antisense) strand. Used for translation of protein. |
| 3′–CGCUAUAGCGUUU–5′ | mRNA antisense transcript | RNA strand transcribed from the Crick (non-template/sense) strand. |
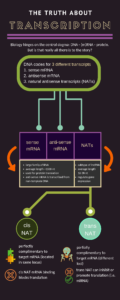
The strand complementary to the Watson strand is the—you guessed it—Crick strand, which can also code for mRNA. The mRNA from the Crick strand will be read from 3’ to 5’ as well. So, you cannot just ignore the ‘minor’ Crick strand. According to Zhao S et al (1), among the over 57,800 annotated genes in the human genome, there are approximately 19% (about 11,000) of overlapping genes transcribed from the opposite strands. One example of a human gene that is read from both strands of DNA is GnRH (2). To determine from which strand your RNA transcript originates, you must use strand-specific RNA-sequencing in transcriptome profiling. This will allow for you to identify non-conventional transcripts such as antisense non-coding transcripts; they are often located on the DNA strand opposite to the origin of mRNA they regulate (1).
Antisense RNA
But mRNA is not the only important transcript type that can be read from DNA. So-called natural antisense transcripts (NATs) are complementary to other RNA transcripts and are also read from both DNA strands (3).NATs have several regulatory roles, mostly in epigenetics.
NATs have several regulatory roles, mostly in epigenetics. For example, NATs epigenetically regulate RNA interference, alternative splicing, and genomic imprinting. NATs are grouped as cis- or trans-acting. Cis-acting NATs are located in the same locus as the mRNA they regulate. This RNA is transcribed from the strand complementary to the mRNA. The corresponding NAT and mRNA have the propensity to pair with each other—just as antiparallel DNA strands bind to each other. The mRNA–NAT binding blocks translation. Cis-acting NATs are found in both prokaryotic and eukaryotic organisms.
On the other hand, trans-NATs are transcribed from a location other than their targets and are usually only partially complementary to their targets. They can target several transcripts via incomplete complementarity. For example, microRNAs (miRNAs) are a type of trans-NAT that imperfectly pair with sequences in the 3’ untranslated region (UTR) of distantly located mRNAs to either inhibit or promote translation (4).
Methods of Detection
One can easily predict an mRNA from the DNA sequence. The mRNA has specific features, like a transcription and translation start, translation stop, polyadenylation sequence, and so on. On the other hand, NATs are much shorter than the typical mRNA (usually just tens of nucleotides) and are located almost anywhere (including introns). Several computational methods for NAT prediction are available, all of which rely on the NAT conservation across species and clustering. However, they have an increased uncertainty, which makes confirmation of NATs by transcriptome analysis especially important. For example, Dongliang Yu and co-workers have proposed an integrated (and freely available) method of NAT prediction from transcriptomes in plants and animals (5).
The myth of the scarcity of functional genes in higher organisms genomes comes from the definition of the gene as a protein-producing stretch of the DNA. In fact, both DNA strands take part in transcription and produce a variety of functional RNA transcripts, of which mRNA are only a tip of the cellular transcriptome iceberg. Therefore, it is important to make sure that you know from which strand your transcripts originate and to consider NATs as a default, not as an exception.
One way to ensure you know the strandedness of your transcripts is to use a reliable kit designed specifically for the preparation of cDNA libraries for strand-specific RNA sequencing. Enter the InvitrogenTM CollibriTM Stranded RNA Library Prep Kit for IlluminaTM with CollibriTM Human/Mouse/Rat rRNA Depletion Kit by Invitrogen (Thermo Fisher Scientific)! The stand-out star of this kit is the Adaptor Mix, which is a set of RNA/DNA oligonucleotides with single-stranded degenerate sequence at one end and a defined sequence at the other end. These oligonucleotides hybridize with the RNA sample (following rRNA depletion and fragmentation), and are then ligated with a ligation enzyme mix. And voilà – a simple and effective way to keep track of your RNA strandedness!
For more information visit our website (Invitrogen Collibri) and click below to download our free infographic!
Sources:
- Zhao S., et al. (2015) Comparison of stranded and non-stranded RNA-seq transcriptome profiling and investigation of gene overlap. BMC Genomics. 2015; 16(1): 675. doi: 1186/s12864-015-1876-7
- Cartwright R. A. and Graur D. (2011) The multiple personalities of Watson and Crick strands. Biol Direct. 2011; 6:7. doi: 1186/1745-6150-6-7
- Osato N., et al. (2007). Transcriptional Interferences in cis Natural Antisense Transcripts of Humans and Mice. Genetics. 176 (12): 1299–306. doi:1534/genetics.106.069484.
- Carmichael G.G. (2003). “Antisense starts making more sense”. Nat Biotechnol. 21 (4): 371–2. doi:1038/nbt0403-371
- Yu D.et al. (2016) NATpipe: an integrative pipeline for systematical discovery of natural antisense transcripts (NATs) and phase-distributed nat-siRNAs from de novo assembled transcriptomes. Scientific Reports volume 6, Article number: 21666 (2016) doi: 1038/srep21666.