The luciferase reporter assay is commonly used as a tool to study gene expression at the transcriptional level. It is widely used because it is convenient, relatively inexpensive, and gives quantitative measurements instantaneously.
It has broad applications across various fields of cell and molecular biology—wherever you want to measure or track the expression of a cloned gene.
As with many assays and kits, many people who use the luciferase reporter assay don’t take the time to fully understand how it works. But understanding the basics of the techniques you are using is essential for troubleshooting when you hit a problem, and for convincing your thesis examiners that you know what you are talking about!
So let’s dive into the inner workings of the luciferase assay.
The Reaction at the Heart of the Luciferase Reporter Assay
Luciferases make up a class of oxidative enzymes found in several species that enable the organisms that express them to “bioluminesce,” or emit light. The most famous one of these enzymes is the firefly luciferase. Fireflies are able to emit light via a chemical reaction in which luciferin is converted to oxyluciferin by the luciferase enzyme. Some of the energy released by this reaction is in the form of light.
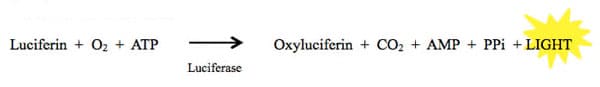
This reaction is highly energetically efficient, meaning nearly all the energy put into the reaction is rapidly converted to light. This makes it extremely sensitive, which is great for a reporter assay!
As an interesting aside… bioluminescence serves a variety of different purposes in nature. Some of these functions include communication, finding a mating partner, finding food, camouflage, and self-defense. In insects, it is thought to play an additional role in oxygen detoxification, a function that presumably evolved more recently. But I digress… back to the assay…
Carrying Out the Assay
There are several commercially available luciferase reporter assay kits comprising expression vectors that contain the luciferase reporter gene (or a variation of it) and the reagents necessary for the reaction to occur.
To perform the reporter assay, you clone the regulatory region of your gene-of-interest (X) upstream of the luciferase gene in one of these expression vectors, introduce that resulting vector DNA into cells, and let the cells grow for a period of time to allow transcription and translation to occur.
You then collect the cells, break them open to release all the proteins (including the luciferase), add luciferin and all the necessary cofactors, and measure the enzymatic activity using a luminometer (an instrument that measures light emission from samples and gives you a quantitative reading).
Since X controls the expression of the luciferase reporter gene, the luciferase activity can be directly correlated with the activity of X.
What Information Does a Luciferase Assay Tell you?
The luciferase reporter assay allows you to study the regulatory control of a gene of interest. The presence of light means that luciferase was transcribed, and therefore the regulatory region was active. This can allow you to determine if a particular protein is involved in the expression of your gene of interest or if the regulatory region of your gene of interest is active under certain conditions.
Disadvantages of Using Luciferase as a Reporter
While the luciferase reporter assay has some great benefits, it also comes with some disadvantages:
- Cells usually need to be lysed before transcription can be quantified (although there are some ways to measure luciferase activity in living cells).
- Luciferase requires ATP to convert the substrate. This means that the metabolic state of the cell can influence the result and introduce bias into the results.[1] However, you can overcome this using ATP-independent luciferases.
- Leaky promoters can sometimes result in an increase in background signal, but unstable reporters have been created that are quickly degraded to help overcome this issue.
The Benefits of Using Luciferase to Study Gene Expression
- This reporter assay can be used to study gene expression as well as other cellular components and events that are involved in gene regulation.
- Its extreme sensitivity allows quantification of even small changes in transcription, and the availability of results within minutes of completing your experiment makes it even more appealing.
- Compared to fluorescent reporters, luciferase has a low background signal.
All in all, the luciferase reporter assay may be just the thing you need to shed some light on your project!
Originally published 26 August 2013. Reviewed and updated, September 2021.
References
- Hakkila K, et al. Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal. Biochem. 2002. 301;235–242.