Alu sequences are repetitive DNA sequences that are widely dispersed within the human genome. The name Alu derives from restriction enzyme site, Alu I, within the repetitive sequence. In human, Alu sequences represent about 6-13% of genomic DNA. These “junk DNAs” are not as useless as one might think. Many studies have suggested that these “junk DNAs” play an important role in genome maintainence and regulation. But they can be used for other things as well.
Today, I would like to tell you an interesting method to quantify the number of integrated Human Immunodeficiency Virus (HIV) genome copies using Alu-PCR. But first, I‘ll tell you how scientists and clinicians have been working with Alu sequences.
Genomic fingerprinting using Alu repetitive sequences
So, what can we do with unique sequences like Alu? We can take advantage of the fact that the location and copy number of the Alu repeats are unique in each person. Therefore, Alu sequences kind of work like a genetic fingerprint of an individual. For this reason, Alu PCR is a powerful technique in forensic science for DNA fingerprinting an individual.
In this modern age, it is performed easily by amplifying regions that are flanked by Alu-specific primers and comparing the genetic polymorphisms between different individuals. In most cases, you should expect the number and the size of the amplified PCR product to be different between each person.
What does ALU sequence have to do with the HIV genome?
One of the coolest features about Alu sequences is that they can be used as internal tags. Scientists use Alu sequences as markers to count the copies of integrated HIV genomes.
HIV is a retrovirus. As such, it reverse transcribes its RNA genome back to DNA and then integrates into the host genome. The insertion location is random, resulting in HIV footprints are all over the host genome.
To devise a strategy to eliminate HIV genomes that are hidden, it is important to identify integrated HIV genomes. In addition, knowing the integrated HIV copy number can help scientists estimate the size of the latent HIV populations and ultimately aid in the design of new anti-HIV compounds. This is where Alu-PCR steps in.
Alu-gag nested PCR coupled with real time PCR for quantitation
Unlike the previous Alu-PCR protocols, the newest method, Alu–gag nested PCR, incorporates 2 steps of PCR to increase the sensitivity and make the assay quantifiable.
Here are the steps:
- Isolate and purify genomic DNA from infected cells.
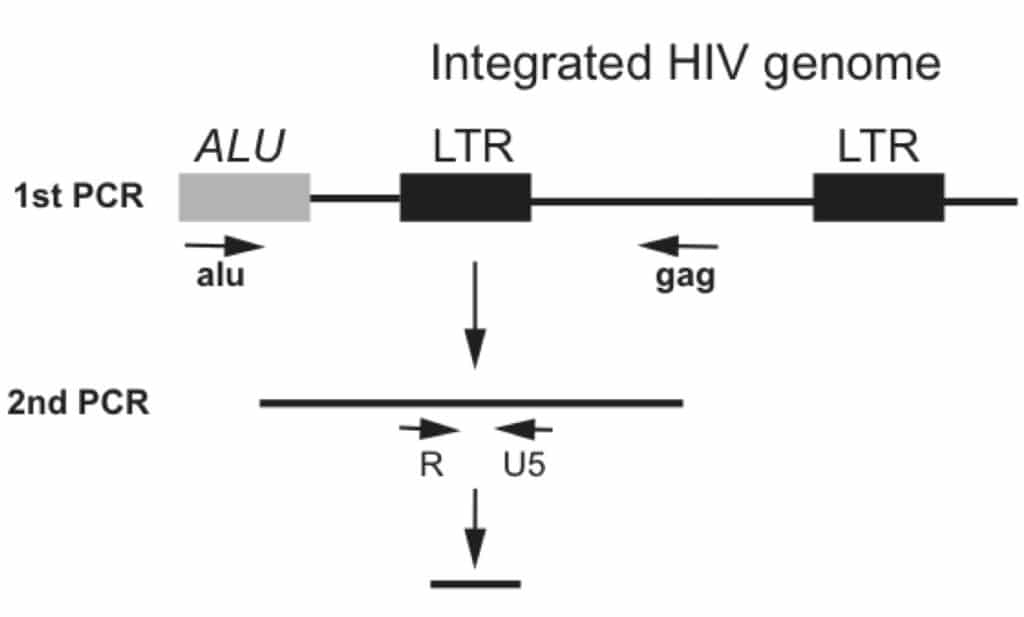
- Perform the first amplification using the primers shown in Figure 1. The primers flank the Alu sequence and the HIV structural gene gag to amplify the gene sequence (Figure 1). Only the integrated form of HIV genomic copies will be amplified exponentially. Un-infected genomic DNA will also be amplified exponentially in this step, but it will not be amplified in the 2nd step in the nested PCR. Finally, un-integrated HIV DNA will only be amplified linearly.
- In the second round of PCR use the forward primer R and reverse primer U5, which flank the viral LTR region (Figure 1). In addition, incorporate a Beckman probe for real-time PCR to quantitate the HIV integration copy numbers. Primer sequences are shown in the table1.
Primer Sequences | |
Alu (forward primer) | 5′ GCC TCC CAA AGT GCT GGG ATT ACA G-3′ |
HIV gag (reverse primer) | 5′ GTT CCT GCT ATG TCA CTT CC-3′ |
R (forward primer) | 5′ -TTA AGC CTC AAT AAA GCT TGC C-3′ |
U5 (reverse primer) | 5′ -GTT CGG GCG CCA CTG CTA GA-3′ |
Potential Limitations
There have been concerns about whether the distance between the Alu sequence and the random integration of HIV genome could lead to under-estimation of the number of integrated copies. However, although it has recently been shown that there is a strong preference for HIV to insert its genome near the Alu sequence, there is no indication that the site of integration has major effects on the performance of Alu–gag PCR.
Although we are fortunate to have drugs available that keep the level of HIV replication in check, understanding the locations of the “hideout” virus will be another step forward in elimination of HIV. Alu-gag PCR is a tool designed to address such issue. Alu-gag PCR is an efficient and robust method to monitor the number of integrated HIV genome copies.
References
- Brady T, et. al. 2013. Quantitation of HIV DNA integration: effects of differential integration site distributions on Alu-PCR assays. Journal of Virological Methods. 189(1): 53–57. doi:https://doi.org/10.1016/j.jviromet.2013.01.004.







