When you’re new to molecular biology, its sheer scope can be a little overwhelming. Many students just starting out in the field have the same questions I did when I first started:
- How do all the different techniques fit together?
- How do I choose the right technique?
- Where do I start?
- And the always popular,
- Why isn’t this working? (Although this one is usually accompanied by many fairly inventive expletives)
It can be a bit of a challenge to tie in all that theory you learnt as an undergrad into an actual workflow, especially if you have only ever used these techniques in isolation (excuse the pun). Worse yet is when your project coordinator suggests a technique and assumes you know all the important steps in-between.
The more experienced hands in your lab know that molecular biology is rarely just a journey from A to B, but it can come as quite a surprise for those of us who haven’t been on that particular expedition. Often a single technique is made up of many smaller steps that may each need their own tips, tricks and optimizations.
As a result, I’ve constructed this short workflow as an introduction to genomic molecular techniques. It is by no means an exhaustive molecular biology guide (I have omitted the whole protein side of things for the sake of simplicity- this is an introduction after all), but it is enough to point you in the direction you need to go (I hope):
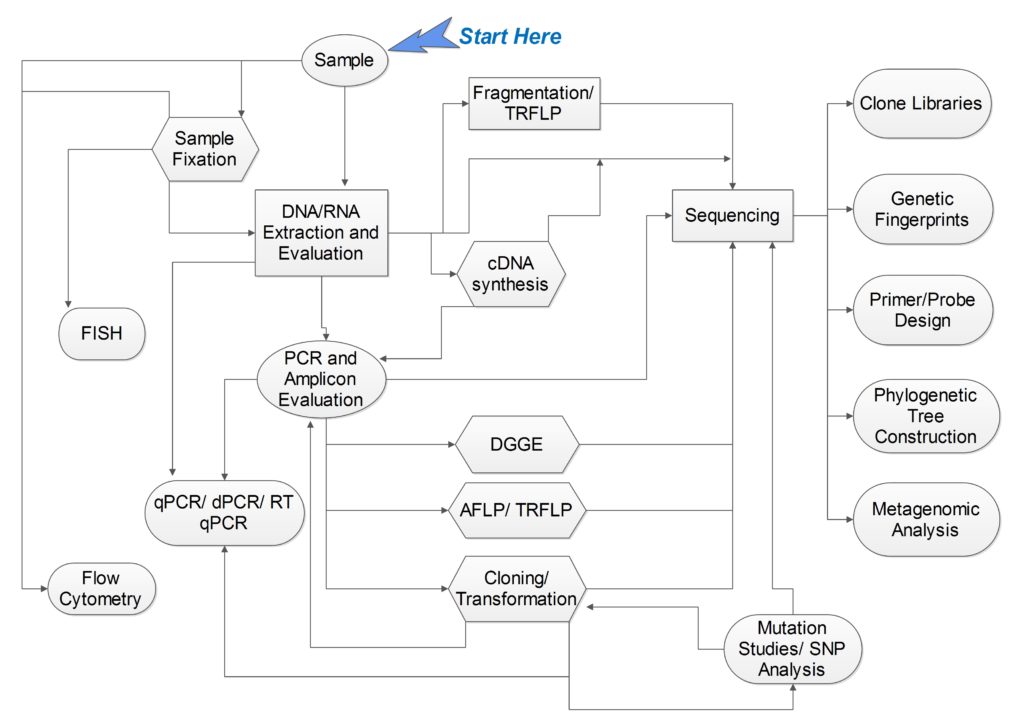
Nucleic Acid Extraction
Your nucleic acid extraction is the first phase of any molecular biology attempt and the efficiency of your extraction will affect all downstream molecular applications. Remember that not all samples respond to treatments in the same way, and it’s always a good idea to understand the nature of your sample in order to choose the best possible method for your sample type. Luckily, Bite Size Bio has a whole channel (which I highly recommend) dedicated to nucleic acid purification to help optimize any type of extraction problem.
PCR
All roads lead to Rome as they say. Well in molecular biology, all roads lead to PCR- and away from PCR- and back to PCR again. Basically, PCR is the central technique in all of molecular biology, and it is one that you ought to master (The BiteSize Bio Guide to PCR is super helpful and definitely worth a place of pride on any respectable scientists bookshelf). And with so many variations to PCR, there will always be one suitable for your application. Here are also some other worthwhile links to check out if you really want to become the PCR guru in your lab:
- https://bitesizebio.com/11004/pcr-is-30-heres-how-it-has-rocked-the-world/
- https://bitesizebio.com/9331/tips-for-amplifying-difficult-pcr-substrates/
- https://bitesizebio.com/4074/the-pcr-controls-you-must-use/
Cloning and Transformation
The importance of transformation cannot be underestimated with regard to protein expression studies, but it is also an invaluable technique in the context of pure genomics. Transformation of competent bacterial cells is most often associated with purification of PCR amplicons or the construction of a standard curve for use in qPCR/ RT-qPCR.
Sequencing
The ultimate goal of course, is to sequence your hard won genomic data. There are many types of sequencing methods currently available, and the rapidly decreasing cost of Next Generation Sequencing (NGS) are allowing us to obtain vast amounts of useful genomic information. Ironically, sequencing represents both the end of your molecular biology journey and often the start of a whole new one.
I hope this roadmap demystifies the workflow of molecular biology- even if only by a little. Sure there are loads more techniques that I haven’t expanded on (I’m sneakily saving FISH and Flow Cytometry for the next article), but these represent your basics, and once understood will have you creating your own protocols like a boss.
And why wait until you get a degree in molecular biology before trying these- knowing molecular biology techniques will only enhance your skills as a life scientist, so take the plunge- it’s definitely a journey worth going on.
So have I missed any of the important detours? Comment below.





