Hydrolysis probes, commonly also referred to as TaqmanTM is a very popular chemistry for real-time PCR. In this article I will compare hydrolysis probes with PlexorTM. But first, a quick overview of hydrolysis probes.
Hydrolysis probes, an overview
Hydrolysis probes are a popular detection chemistry for monitoring sequence-specific amplification in RTPCR. Just like with SYBR Green dye, signal detection is achieved through monitoring an increase in fluorescence as the reaction proceeds. But, the fluorescent signal in TaqmanTM chemistry is dependent on probe hydrolysis, rather than hybridization, hence the name “hydrolysis probes”! (1).
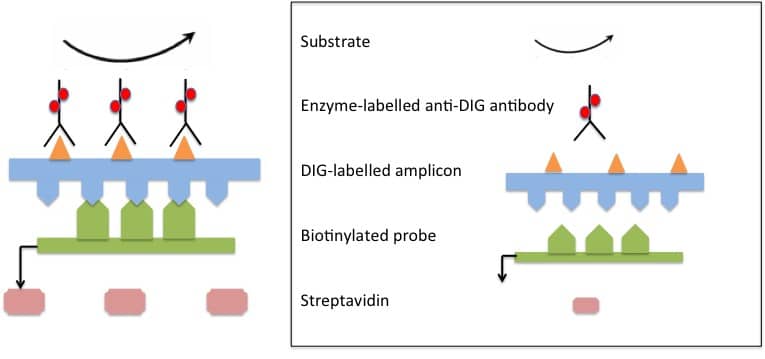
In the hydrolysis probes set up, there are two primers and a probe. The probe, also designed complementary to the target, contains the fluor and quencher on either end (see the figure below).
During the amplification process, the probe binds to the specific target sequence during the annealing step. Because of the proximity between the donor (fluor) and acceptor (quencher) on the probe, there is no fluorescence. During the extension step, the 5′-3′ exo activity of the polymerase, hydrolyses the probe relieving the fluor from quenching effects and fluorescence is read by the detector.
So, how does this compare with Plexor?
Signal Specificity & Multiplexing
Sequence specific amplification in general produces greater signal specificity than intercalator-dye based methods. So, both hydrolysis probes and PlexorTM can be considered to have equivalent signal specificity.
Both technologies are amenable to multiplexing options by using probes or primers with different fluorescent labels for each target. However, optimization of multiple primers is done more easily with PlexorTM since there are no extra probes to design and optimize.
Melt Curve Analysis
Hydrolysis probes show reaction profiles only for sequence specific amplification, so extent of primer-dimer and non-specific product formation, both of which can affect amplification efficiency of the target, remains unknown. The ability to follow up with a melt curve analysis after real-time quantification in the same reaction set-up is therefore a big advantage.
In PlexorTM melt curve analysis can be programmed as part of the cycling protocol. During data analysis using the in-built software, melt profiles for specific & non-specific products and PDs can be easily differentiated based on the Tm.
Signal Sensitivity
The fluorescent signal obtained when using hydrolysis is cumulative. This means that the fluorescent signal of the cleaved probe from one cycle contributes towards signal in the next cycle of the same reaction. Because of this, concentrations of targets that are very low or limiting may not be correctly assessed.
With PlexorTM, the signal (quenching of fluorescence) is a direct measure of product concentration as quenching occurs at every cycle all over again. The signal is therefore more specific (2).
Ease of use
PlexorTM chemistry is definitely easy to use and optimize. Designing good primers is a requirement for any PCR and with PlexorTM the primer design software (see here) makes that job easier. So, ordering primers is all it takes to get the chemistry running.
With the probe chemistry, Tms between probes and primers will have to be optimized to allow efficient annealing during the reaction. If the Tm of the probe is off, the signal obtained will not be accurate.
Finally…..
Some of the disadvantages of PlexorTM apply to all technologies that use fluorescent dyes for signal detection such as instrument capability, dye compatibility, storage and shelf-life of labelled nucleotides etc. Other PlexorTM specific disadvantages are listed in my previous article:
1. Iso-dG is part of the dNTP mix. It has to be purchased from Promega and cannot be used as part of any non-Plexor PCR reaction.
2. As with any fluorescence technology, the reagents have to be protected from light. Since fluorescence labels are known to degrade with time, the cost of reagents can be expensive depending on throughput.
3. The absorption maximum of Dabcyl occurs at 474nm which falls below the emission wavelength of commonly used fluorophores such as FAM, TET and JOE. This means that quenching efficiency of Dabcyl is not 100%. It is therefore important to exercise good care in choosing the right fluorophore .
It was brought to my attention that pre-designed assays are also a possibility with PlexorTM technology. An example of this is Promega’s PlexorTM HY system that quantifies both total human DNA and male DNA from the same reaction.
The possibilities with PlexorTM seem endless. Do you think so? Why would you or would you not use PlexorTM chemistry?
References:
- Francois Ferre. 1997. Chapter” Fluorescence Monitoring of Rapid cycle PCR for Quantification. Gene Quantification. ISBN 0-8176-3945-4. ISBN 3-7643-3945-4 p. 133.
- Douglas. R. Storts. A Comparison of PlexorTM and 5’Nuclease Assay Chemistries