Around and around the cell cycle goes, where it stops, nobody knows.
Unless you have the right tools to analyze DNA content, that is.
The DNA markers propidium iodide, Hoechst and DAPI are commonly used in flow cytometry to analyse a cell’s DNA content. Although they are simple to use, they do have disadvantages.
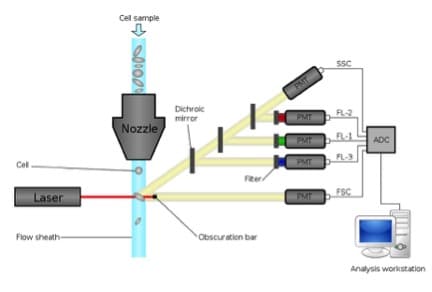
Figure 1 shows a typical DNA profile analyzed using propidium iodide. You can easily see defined G1 and G2 peaks and S phase cells between the two. However, this plot only offers a snapshot of the cells at one time-point – you can’t determine if the cells in S phase are actually cycling and also have no idea of the kinetics of the cycle; you can’t tell how long cells take to pass through each stage of the cell cycle. To address this, cytometrists have developed several approaches.
How BrdU works
One of the more common ways to assess cell proliferation is to use a thymidine analog, which is taken up only by cycling cells.
The first thymidine analog that was used was bromodeoxyuridine (BrdU). BrdU is easy to use – you can add BrdU to cell cultures, into an animal’s drinking water or inject it directly into human tumours.
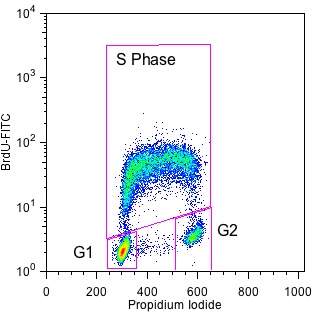
The principle is simple – when a cell replicates DNA, BrdU is incorporated in place of thymidine. Incorporated BrdU can then be labeled with a specific antibody and detected by flow cytometry in conjunction with propidium iodide staining (Figure 2).
The detection of incorporated BrdU is quite straightforward but as the BrdU is detected via an antibody, the DNA must be unwound to allow access of the antibody to its target (BrdU). There are standard ways of doing this using either strong acid or alkali, heat or enzymatic treatment.
What can BrdU do for you?
Using BrdU, you can more easily identify cells in G1, S and G2. This allows you to better define the cells in each phase and answer questions about cell cycle kinetics.
For example, if you want to know how many cells are cycling at a given time, you can expose cells for a short time (30 minutes) to BrdU and compare the cycling in different cell types or possibly the same cell type following different treatments.
Or perhaps you want to know if there are quiescent, non-cycling cells in your culture. This is easily answered using BrdU incorporation. Expose your cultures to BrdU for a long time, 24 hours for example. Any cells that remain BrdU-negative never entered the cell cycle and did not cycle during that timeframe.
If you are interested in how long it takes cells to traverse different stages of the cell cycle, you can label with BrdU in a “pulse-chase” format. Add BrdU to a culture for 15 minutes (pulse) and then replace the medium with one that contains no BrdU (chase). Then take samples at different timepoints after the addition of the chase medium. You can then follow the cells that were in S phase at the beginning of the experiment (BrdU positive) and determine where they end up. So, for example, when you see labelled cells re-appearing in G1, you know that they have taken that long t pass through G2 and mitosis.
Making it better
As mentioned above, BrdU can only be detected by anti-BrdU antibodies after treating the cells to unwind the DNA. Unfortunately, several of these treatments are harsh and are not compatible with labelling of other antigens.
To overcome this, another thymidine analog, ethynyl-deoxyuridine (EdU) has been introduced. The protocol uses a copper catalysed reaction to add a fluorescent azide to an alkyne group on the incorporated EdU. As this doesn’t involve unwinding of DNA there is a greater variety of simultaneous antigen detection available.
You only get to see one round 1
Although thymidine analogs can tell you a lot about the cell cycle, they are only capable of following cells through one and a half divisions. After that, the synchronicity of labelled cells begins to fail.
You can get around this by using alternative methods (such as the Hoechst-BrdU quenching method) but the technique is technically difficult and also requires a cytometer equipped with a UV laser.
If you want to look at multiple rounds of cell division, stay tuned for our Cell Proliferation Round 2 post, which will tell you about the dye dilution method for cell cycle analysis.