As biological catalysts, enzymes transform target substrates into products. Enzyme kinetics is the rate of that transformation. By understanding how an enzyme’s behavior is affected, you can figure out how it functions in physiology or fails to function in disease.
Now it gets complicated…
What Affects an Enzyme’s Kinetics?
In the first place, most enzymes are tightly regulated: they won’t be active or synthesized until needed. Furthermore, many enzymes need small molecule cofactors to do part of their reactions. These cofactors can be metals, like zinc or iron, or organic coenzymes (e.g. NADPH).
The reaction rate also depends on the environment. Enzymes function at an optimal pH; when the environmental pH is different, the enzyme loses structural stability and its reaction rate decreases. Enzymes also have optimal temperatures, and their rates accelerate as temperatures increase to 50°C, but decrease abruptly as the enzyme denatures around 55-60°C. The exceptions to this are enzymes found within thermophilic bacteria, which operate maximally at a higher temperature.
An enzyme’s kinetics can also be controlled by inhibitors. Inhibitors reduce activity by either blocking the interaction between the enzyme and substrate or altering the substrate’s transformation rate. Many pesticides (e.g. chlorpyrifos) and drugs (e.g. ACE inhibitors) are specific enzyme inhibitors.
How Do We Figure Out Enzyme Kinetics?
To determine kinetics, we analyze an enzyme’s activity – the number of reactions it manages over time. After setting the length of time and amount of enzyme and substrate, it’s possible to determine the velocity – the reaction rate – by measuring product appearance (or substrate disappearance) at different times. By plotting the amount of product versus time, we can determine velocity from the linear portion of the curve.
You Knew It Was Coming: Michaelis-Menten Kinetics!
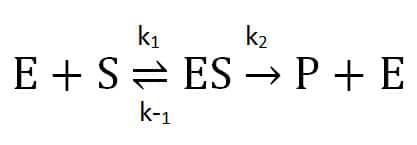
Is it really that simple? Perhaps – if you are fortunate enough to study a classic Michaelis-Menten enzyme! The Michaelis-Menten theory describes a reaction involving one substrate (S), enzyme (E), an intermediate enzyme-substrate complex (ES), and a product (P) and regenerated enzyme. This assumes that when the enzyme complexes with the substrate, it either dissociates into unchanged substrate and enzyme or proceeds irreversibly forward to product.
The second step’s rate (k2) is the enzyme’s turnover for enzyme-substrate converting to product. We get the Michaelis constant, Km, from the ratio of the reactions affecting the enzyme-substrate complex: its formation (k1), dissociation (k-1), and progress towards product (k2). A lower Km value indicates that an enzyme has a higher affinity for a substrate.
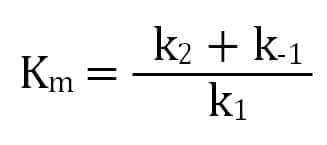
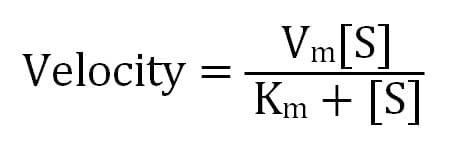
Ultimately, a Michaelis-Menten enzyme’s velocity depends on its Km, substrate concentration, and maximum possible velocity (Vm), which occurs when there is much more substrate than enzyme. This means that when the substrate concentration equals Km, the enzyme’s velocity is half of its Vm.
How Do Enzyme Inhibitors Affect Michaelis-Menten Kinetics?
If an inhibitor acts irreversibly, it destroys an enzyme’s ability to transform its substrate, usually by covalently modifying the enzyme. The effects of an irreversible inhibitor will be observed in a decreased reaction velocity over time.
However, if an inhibitor is reversible, you can measure its effects on Vm and Km. These effects depend on whether the inhibitor competitively blocks the enzyme’s active site (Vm stays the same, Km increases), noncompetitively binds outside the active site (Vm decreases, Km stays the same), or noncompetitively binds to the enzyme-substrate complex (Vm and Km both decrease).
Is There Anything Other Than Michaelis-Menten Kinetics?
Not all enzymes follow Michaelis-Menten kinetics. What if the product transforms into substrate, inhibit its own formation, or the enzyme’s behavior depends on substrate concentration?
For example, allosteric enzymes – which have at least two substrate sites – do not follow Michaelis-Menten. When the first substrate binds to the enzyme it alters the enzyme’s affinity for the second enzyme, often via a conformational change. To learn more about allosteric kinetics, I would recommend looking into the resources below.
Putting It Into Practice
Equations that explain enzyme kinetics are all well and good, but you need to get those numbers to fill in the variables. There are several different techniques for measuring the binding and dissociation of proteins that can be applied towards enzyme kinetics.
Here’s a quick spotlight on a couple:
Förster Resonance Energy Transfer (FRET)
FRET is a sensitive technique for studying the interaction of certain types of enzymes, for example hydrolytic enzymes like proteases. In FRET, interacting proteins are labeled with fluorescent donor and acceptor molecules. When two binding partners are closely opposed, the donor label transfers energy to the acceptor molecule causing it to fluoresce. You can measure absolute fluorescence levels from both the donor and acceptor to quantify kinetic parameters.
Surface Plasmon Resonance (SPR)
If you want a highly quantititave assay that doesn’t require labeling of proteins, then turn to a SPR assay. An SPR assay uses optical measurements to quantify binding interactions between molecules. One molecule is bound to a specialized sensor chip while the other is flowed over the sensor surface. An interaction between the molecules causes changes in the refractive index of light that is passed through the sensor.
This was just a quick primer on the complicated world of enzyme kinetics. Although it may not be for the faint of heart, kinetics is fascinating!
Resources:
- Atkins WM. Michaelis-Menten Kinetics and Briggs-Haldane Kinetics. https://depts.washington.edu/wmatkins/kinetics/michaelis-menten.html.
- Costa LG. Toxic Effects of Pesticides. In: Klaassen CD, ed. Casarett and Doull’s Toxicology: The Basic Science of Poisons. 7th ed. New York, NY: The McGraw-Hill Companies; 2006:883-930.
- Robyt JF, White BJ. Biochemical Techniques Theory and Practice. Prospect Heights, IL: Waveland Press, Inc.; 1990:291-320.
- Voet D, Voet J. Biochemistry. 3rd ed. Hoboken, NJ: John Wiley & Sons; 2004:459-546.







