RNA sequencing (Wang 2009) is rapidly replacing gene expression microarrays in many labs. RNA-seq lets you quantify, discover and profile RNAs. For this technique, mRNA (and other RNAs) are first converted to cDNA. The cDNA is then used as the input for a next-generation sequencing library preparation. In this article, I’ll give a brief review of RNA-seq and introduce the major methods being used today.
Why Is RNA-Seq “Better” Than Microarrays?
There are several advantages RNA-seq has over microarrays:
With RNA-seq you can interrogate more than just differential gene expression. Although there are microarrays available for exon-level and microRNA analysis, most users are still interested in basic, probably 3’ biased, differential gene expression. With RNA-seq you can look at coding and non-coding RNA, at splicing and allele specific expression, and possibly soon at full-length cDNA sequences, eliminating the need to infer or assemble isoforms.
Microarrays are also biased, as we have to decide what content to place on the array. Since RNA-seq does not use probes or primers, the data suffer from much lower biases (although I do not mean to say RNA-seq has none).
RNA-seq provides digital data in the form of aligned read-counts, resulting in a very wide dynamic range, improving the sensitivity of detection for rare transcripts.
It is also very cost-competitive to microarrays, as today, between 6-30 samples can be multiplexed in a single Illumina sequencing lane.
Lastly, you can reanalyze an RNA-seq dataset as more information about the transcriptome becomes available. If a paper is published showing an interesting splice-variant in a similar system to the one you work on, then you might want to go back and look at that splicing in your samples; and you’d already have the data to do so.
How Does RNA-Seq Work?
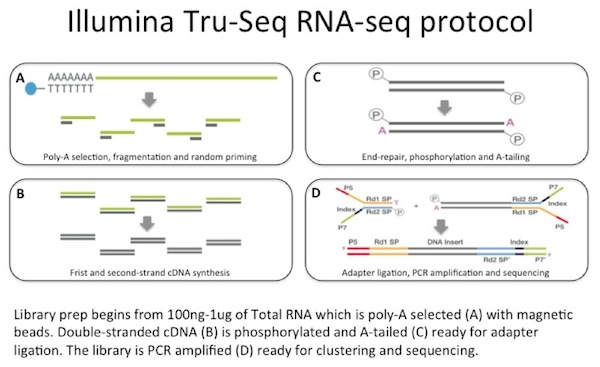
There are many methods for performing an RNA-seq experiment. In fact, the techniques are evolving so rapidly it can be difficult to decide which one to use. A basic choice is between 1) random-primed cDNA synthesis from double-stranded cDNA or 2) RNA-ligation methods (reviewed and compared in Levin 2010). Most people use the first method and then need to make a further choice between a strand-specific protocol and one that is not. The method used most in my lab is Illumina’s TruSeq RNA-seq, which is a random-primed cDNA synthesis non-strand-specific protocol.
Once you have a sequencing library, it is sequenced to a specified depth, which is dependent on what you want to do with the data. These reads are aligned to the genome or transcriptome and are counted to determine differential gene expression or further analyzed to determine splicing and isoform expression. Most people are sequencing RNA using paired-end 50-100bp methods. The exception is microRNA sequencing, as this only requires single-end 36bp sequencing in most cases.
Our RNA-Seq Method
We use between 100 ng to 1 µg of total RNA as the input to an mRNA capture with oligo-dT coated magnetic beads. The mRNA is fragmented, and then a random-primed cDNA synthesis is performed. The resulting double-strand cDNA is used as the input to a standard Illumina library prep which includes end-repair, adapter ligation and PCR amplification to give you a library ready for sequencing.
Why Bother With Strand Information?
There has been a lot of discussion about anti-sense transcription and its biological relevance. If you are interested in simple differential gene expression, then strand information will not add much to your experiment, but will make your protocol more complex. Having said that, you can perform the most widely adopted method without too much extra effort. To do this, during 2nd strand cDNA synthesis, use uracil for incorporation instead of thymine. Follow the Illumina library prep as normal, but after adapter ligation and before PCR amplification add uracil-DNA glycosylase to degrade the 2ndstrand. This results in all reads starting in the same orientation so you can determine which strand was being transcribed in your sample.
What Can You Actually Do With RNA-Seq?
RNA-seq is a powerful and versatile tool published widely over the last few years. I have picked a couple of my favorites (some from work performed in the core facility I manage) to illustrate what you can do with RNA-sequencing.
- Jabbari, et al. used RNA-seq to investigate psoriasis and find new genes for functional analysis. They compared their RNA-seq data to published array studies and found 1700 new candidates. These were validated by qPCR, and comparison to functional databases for psoriasis supported their role in pathogenesis.
- Kutter, et al. used RNA-seq in a study looking at the conservation of RNA Polymerase III binding in mammals to validate expression of genes occupied by Pol III as assayed by ChIP-seq.
- Mercer, et al. combined RNA-seq and microarray-based capture to identify and characterize rare transcripts, which are normally undetectable. The targeted transcripts increased in sequence read abundance from 0.21% pre-capture to 80% post capture. They found more than 200 previously unannotated isoforms for almost 50 protein-coding loci, including a new alternative isoform of TP53, which is a very well characterized gene. This suggests that there is still much complexity in the genome and transcriptome to be resolved.
In summary, RNA-seq is still an evolving tool, but is preferable in most instances to microarrays. It is more sensitive, more robust and can be more cost effective. What RNA-seq projects are you now planning for your project?
References:
Wang et al: RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009.