Do you need to brush up on your alternative splicing knowledge? Well, look no further! We’re here to help with our guide to this splicing mechanism. But before we get stuck in, let’s start with some background information.
The Central Dogma of Protein Synthesis
We know that an organism’s genetic information is stored in its genes, the functional subunits of the genome. Genes are arranged in the strands of the DNA double helix in the nucleus of cells.
We also know that this information is transcribed from DNA into a messenger RNA (mRNA) template in a process called transcription. As the name suggests, this mRNA template then acts as a messenger to synthesize proteins in a process called translation.
This is the first part of what is generally known as the central dogma of molecular biology, which was first described by Francis Crick in 1958. [1]
But, as you probably know by now, biology is never that simple. After uncovering the general mechanisms behind protein synthesis, it was later discovered that in viruses, some non-coding mRNA coding sequences, known as introns, need to be removed or “spliced out” from the final mRNA molecule before it can be translated into protein. [2,3]
Removal of these introns leaves only the protein-coding regions, called exons, which must be joined by RNA splicing to produce mature mRNA to allow for the translation of a functional protein. These findings were later extended to other organisms including eukaryotes.
As a quick summary—most genes in higher eukaryotes are transcribed as pre-mRNA, which contains non-coding and coding regions known as introns and exons, respectively.
In a process mediated by the spliceosome, introns are removed while exons remain to give a final mature mRNA sequence.
If you’re anything like me, I always found the terminology here very confusing. I would have expected introns to be kept in the sequence and exons to exit the scene. But that would be too obvious!
Here’s a good mnemonic to help keep you right—exons are expressed in the final mRNA while introns are in the trash (and therefore not in the mature mRNA). OK, let’s get onto the nitty-gritty.
How Was Alternative Splicing Discovered?
RNA splicing was discovered in the 1970s, first in viruses and then in eukaryotes [2,3]. Soon after, scientists discovered alternative patterns of pre-mRNA splicing that produced different mature mRNAs containing various combinations of exons from a single precursor mRNA.
Therefore, alternative splicing, a type of post-transcriptional modification, is the process by which exons or portions of exons or non-coding regions within a pre-mRNA transcript are differentially joined or skipped, resulting in multiple protein isoforms being encoded by a single gene.
As a result, alternative splicing increases the complexity of the proteome that can be generated from the available genome sequences.
The first example of alternative splicing of a cellular gene in eukaryotes was identified in the IgM gene, a member of the immunoglobulin superfamily. [4,5]
This splicing mechanism increases the informational diversity and functional capacity of a gene during post-transcriptional processing and provides an opportunity for gene regulation (Figure 1).
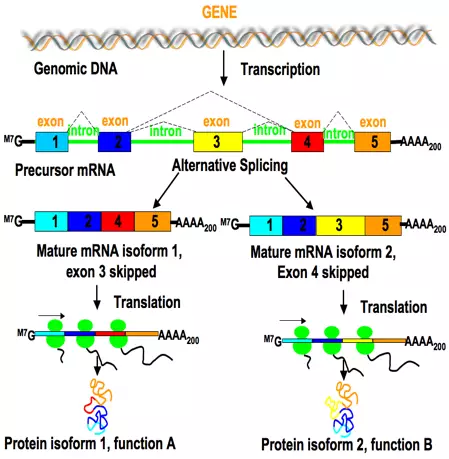
Figure 1. Alternative splicing generates transcriptome diversity and enables gene regulation. It can generate mRNAs that encode proteins with different or even opposite functions. Figure used with permission. [6]
Still with me? Great! So, what’s the point of alternative splicing?
Well, it plays a crucial role in generating biological complexity and proteomic diversity in humans and significantly affects various functions in cellular processes, tissue specificity, developmental states, and disease conditions.
Because it’s so important in many biological mechanisms and is required to regulate normal physiological functions, it is vulnerable to dysregulation.
As a result, alternative splicing is often involved in human disease (e.g. cancer) [7] and ageing. [8]
How Does Alternative Splicing Work?
In molecular biology a cis-acting regulatory element is defined as a region of chromosomal DNA that regulates the transcription or expression of a gene that is found on the same chromosome.
A trans-acting regulatory element, on the other hand, is a protein that binds to a cis-acting element of a specific gene to regulate its transcription. [9]
As a result, cis-acting regulatory elements in pre-mRNA sequences determine which exons are retained and which exons are spliced out.
These elements alter splicing by binding different trans-acting protein factors such as serine–arginine-rich (SR) proteins, which function as splicing facilitators, and heterogeneous nuclear ribonucleoproteins (hnRNPs), which suppress splicing. [10]
Silencing can be inhibited sterically when splicing inhibitors bind to splicing silencers that are found near splicing enhancers.
This prevents the binding of small nuclear ribonucleoproteins (snRNPs) and other activator proteins or prevents spliceosome assembly. [10]
The inclusion or splicing of an alternative exon is therefore determined by combinatorial effects, cellular abundance, and competitive binding between SR activators and hnRNP inhibitors.
Alternative splicing outcomes depend on the stoichiometry and interactions of splicing activators and inhibitors, as well as the steric conformation and accessibility of the splicing sites.
Why Is Alternative Splicing Important?
On average, human transcripts contain approximately nine introns. This relates to several hundred thousand introns across the genome, which take up 25% of the DNA content of each cell.
Importantly, alternative splicing has been associated with approximately 15% of hereditary diseases and cancers in humans. [11]
Advances in high-throughput technologies, including next-generation RNA and DNA sequencing, have facilitated studies of genome-wide alternative splicing.
These studies estimate that >95% of human multi-exon genes express multiple splice isoforms. [7,12,13]
Alternative splicing events are differentially regulated across different tissues and during development, as well as among individuals and populations.
This suggests that individual isoforms may serve specific spatial or temporal roles. [14,15]
CD44: An Example of Alternative Splicing
For example, several alternatively spliced genes are known to be involved in immunity. [16,17]
Studies indicate that alternative splicing of CD44, a protein involved in T-cell homing with 10 variable cassette exons and six distinct protein isoforms, is crucial for T-cell function. [17]
The variable exons of CD44 encode portions of the membrane-proximal extracellular domain of the protein, and the presence of some of the variable exons has been shown to increase the association of CD44 with various proteins. [17]
Isoform expression is activation dependent. This allows naïve T cells to express the smallest CD44 isoform, which lacks all variable exons.
In comparison, activated T cells express multiple CD44 isoforms, thereby suggesting that CD44 alternative splicing is important for activation. [17]
Alternative Splicing Summarized
The scope and role of this vital regulatory mechanism still requires investigation on a genome-wide scale. It is now clear that alternative splicing is more widespread and complex than was initially thought.
In addition, the role of alternative splicing in different human pathologies is also being revealed.
As a result, improving our understanding of the mechanisms behind alternative splicing will present exciting opportunities for therapeutic development.
Check out our companion article for more on what alternative splicing means for your experiments.
References
[1] Crick FH (1958) On protein synthesis. Symp Soc Exp Biol 12:138–63.
[2] Chow LT et al. (1977) An amazing sequence arrangement at the 5? ends of adenovirus 2 messenger RNA. Cell 12:1–8.
[3] Berget SM et al. (1977) Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. PNAS 74:3171–5.
[4] Liu CP et al. (1980) Mapping of heavy chain genes for mouse immunoglobulins M and D. Science 209(4463):1348–53
[5] Early P et al. (1980) An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell 19(4):981–92.
[6] Grigoryev YA (2011) Post-transcriptional Mechanisms of Gene Regulation and Information Control in Immunity. The Scripps Research Institute, La Jolla, CA.
[7] Wang J et al. (2021) Alternative splicing: An important regulatory mechanism in colorectal carcinoma. Mol Carcinog. 60:279–93.
[8] Bhadra M et al. (2020) Alternative splicing in aging and longevity. Hum Genet 139:357–69.
[9] Griffiths AJ et al. (2000) ‘Transcription: an overview of gene regulation in eukaryotes’ in An Introduction to Genetic Analysis, 7th edition. New York: WH Freeman.
[10] Kelemen O et al. (2013) Function of alternative splicing. Gene 514(1):1–30.
[11] Jiang W, Chen L (2020) Alternative splicing: Human disease and quantitative analysis from high-throughput sequencing. Comput Struct Biotechnol J. 19:183–95.
[12] Pan Q et al. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 40(12):1413–15.
[13] Irimia M, Roy SW (2014) Origin of spliceosomal introns and alternative splicing. Cold Spring Harb Perspect Biol 6(6):a016071.
[14] Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463(7280):457–63.
[15] Licatalosi DD, Darnell RB (2010) RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11(1):75–87.
[16] Su Z, Huang D (2021) Alternative splicing of pre-mRNA in the control of immune activity. Genes 12(4):574.
[17] Martinez NM, Lynch KW (2013). Control of alternative splicing in immune responses: Many regulators, many predictions, much still to learn. Immunol Rev 253(1):216–36.
Originally published December 28, 2012. Reviewed and updated by Zandile Nare August 2021.