Site-directed mutagenesis studies can be extremely useful for elucidating the function of a gene or protein, or for creating variants of an enzyme with new and improved functions.
There are now many approaches available for generating site-directed mutants, whatever your purpose. In this post I’ll summarize three techniques that will enable you to produce a wide range of mutations, and point you to some useful resources to help you get those techniques up and running.
Note: All of these approaches are good for cloned, genomic or cDNA templates unless indicated (* means good for cloned templates only, ** means good for gDNA or cDNA only).
Technique 1: PCR, with modified primers
Enjoying this article? Get hard-won lab wisdom like this delivered to your inbox 3x a week.

Join over 65,000 fellow researchers saving time, reducing stress, and seeing their experiments succeed. Unsubscribe anytime.
Next issue goes out tomorrow; don’t miss it.
Description:
This type of site-directed mutagenesis uses PCR primers designed to contain the desired change. The PCR primer sequence simply replaces the original sequence – as long as the changes are minimal enough to allow the primer to anneal to the intended target.
Use for:
- Limited base identity changes at the end of the target sequence
- 5’ or 3’ terminal insertions <100 bases
Technique 2: Primer extension
Description:
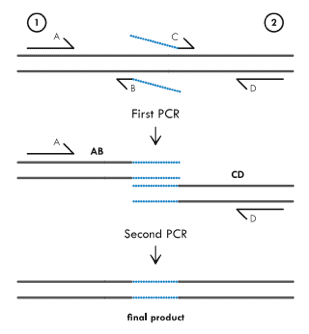
Primer extension uses nested primers to mutate a target region. In the diagram, primers B and C contain the mis-matched sequence to insert bases. The first round of PCR uses primers A-B and C-D to create two products with the mutated sequence.
The second PCR round is where the smart stuff happens and the new sequence is created. Since primers B and C contain complementary sequences, the products from the first round will hybridize after they are denatured following the first PCR cycle. Primers A-D can then be used to amplify the full-length product that contains the desired mutation. Alterations to this method can also create deletions or longer additions.
Use for:
- Limited, non-random base changes internal to the target sequence
- Insertions >100 bases
- Deletions < 50 bases
- Deletions > 50 bases**
Technique 3: Inverse PCR
Description:
Inverse PCR is used for mutating plasmids. This method uses two back-to-back primers to amplify the whole plasmid and the linear product is then ligated back to the circular form. The primer binding regions can be changed by altering the primer sequences to contain the desired mutation. Insertions can be made around the primer binding regions by adding flanking sequences to the primers, and deletions can be made by simply leaving a space between the two primers.
Use for:
- Insertions > 100 bases*
- Deletions < 50 bases*
- Deletions > 50 bases*
Further Reading:
- Kadowaki H et al (1989) Use of polymerase chain reaction catalysed by Taq DNA Polymerase for site-specific mutagenesis Gene 76(1):161-166
- Ho SN et al (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77(1):51-59
- Hemsley A et al (1989) A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 17(16):6545-6551
- Integrated DNA Technologies, 2011. Mutagenesis Application Guide.
You made it to the end—nice work! If you’re the kind of scientist who likes figuring things out without wasting half a day on trial and error, you’ll love our newsletter. Get 3 quick reads a week, packed with hard-won lab wisdom. Join FREE here.