Introduction
Did you know that the idea of using genetic engineering to ameliorate certain human diseases was viewed as ‘science fiction’ only 10 short years ago?
While cell mutagenesis studies and genetic knockout experiments were feasible before genetic engineering, they were not very reliable. Indeed, due to the random and imprecise nature of these older methods, it is required to thoroughly screen the readout to ensure the correct genetic composition.
With the development of new technologies, such as zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), clustered regularly interspaced short palindromic repeats (CRISPR), researchers can alter the genome with more site-improved specificity. This article will focus on the transcription activator-like effector nucleases (TALEN), which is a great technology and used by many labs around the world.
A Bit of Background
Before we can understand how TALENs work, we first need to make sure we cover the basis of DNA repair mechanisms.
Enjoying this article? Get hard-won lab wisdom like this delivered to your inbox 3x a week.

Join over 65,000 fellow researchers saving time, reducing stress, and seeing their experiments succeed. Unsubscribe anytime.
Next issue goes out tomorrow; don’t miss it.
To alter the genetic information in our genome, a double-stranded break (DSB) is required. As you can imagine, having a break or any modification in your genome by natural forces (i.e. ultraviolet (UV) or chemicals (carcinogens) can be deadly to your cells and ultimately your survival as human being. So, when a DSB is made by a nuclease, the emergency DNA repair systems in our cells automatically kicks into gear and try to repair the damaged DNA bases and restore the DNA duplex.
Researchers have taken advantage of this natural process to develop more precise genome editing protocols. By guiding the nuclease to a specific DNA location with a specific sequence, the enzyme will perform DSB at the distinct binding site.
However, as with most biological systems, DNA repair mechanisms are not quite that straight-forward. Indeed, two scenarios could happen depending on the location of the DSB and sequence of the template. The first scenario is non-homologous end-joining, which could result in general frame-shifts, insertions, and deletions. The second scenario is the homologous end-joining, which requires a template of some sort (be it original or artificial), usually result in precise joining based on the template provided. (Ref 1, Figure 1)
How Do TALENs Work?
So what does all of this have to do with TALENs? Everything!
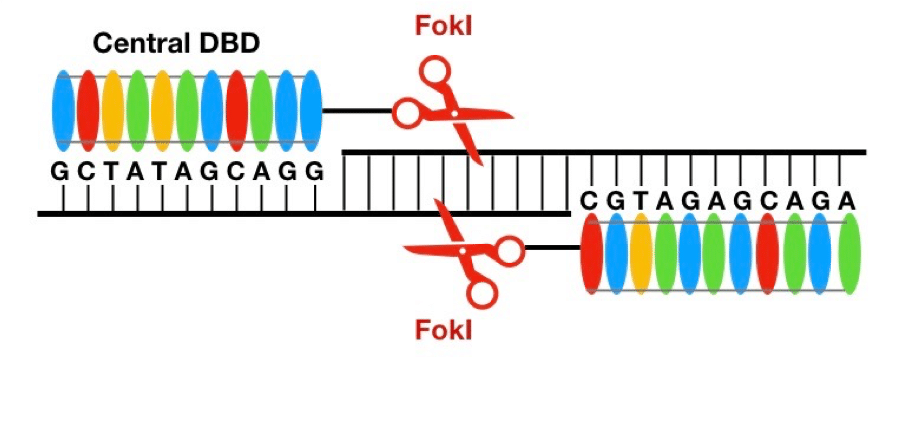
The key technology behind TALEN is the use of transcription activator-like effectors (TALE) to bind a specific stretch of DNA sequence. When coupled with a nuclease such as FokI, the precision of the cutting site is extremely efficient because dimerization is required for FokI nuclease. Therefore, both the forward sequence and the reverse sequence encompassing the target region has to be precise. In addition, you can also pair other functionalities with TALE, such as repressors, etc.
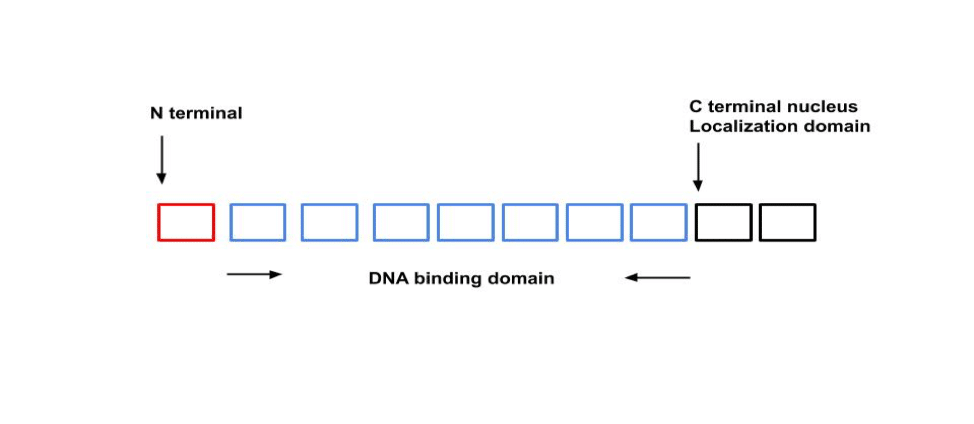
TALEs represent a family of type III effector proteins from Xanthomonas spp., which is a genus of Gram-negative Proteobacteria that is often associated with plant diseases. These clever microbial critters have developed their own ways to control gene expression in plants, primarily through TALEs. TALEs can be divided into 3 sections: 1) N-terminal secretion and translocation signal; 2) a central DNA binding domain (DBD); and 3) C-terminal nuclear-localization domain (Fig. 1).
Investigations into how the DNA-binding domain of TALEs actually worked was researched separately by Boch et al. and Moscou et al. These groups found that the central DNA-binding domain is made up of multiple zones of repeated amino acid (aa) sequences that are 34 unit long (Fig 2). The secret is, within each repeated zone, scientists have discovered that positions 12 and 13 are determinant of the sequence specificity (4). This means that investigators can engineer site-specific sequences for TALEs to bind to; once there, the tethered Fokl endonuclease will perform the DSB (Fig. 2).
Applications
In vitro mutagenesis studies using cell-lines
This is the easiest way to test if your TALEN design works the way it should, and is usually the first step towards more in vivo studies. Introducing your TALEN construct is relatively straight forward, as it involves transfection with cell lines with your plasmid constructs (Ref 3, Figure 3). For example, human embryonic kidney cell line (HEK 293T) and HeLa cell lines are typical choice for verification of gene expression and genotype due to the ease of transfection.
In vivo genome editing
The main reason that researchers embrace the TALEN technology is because its direct-targeting increases the frequency of homologous recombination by several orders of magnitude (1,3). Normally, homologous recombination with a homologue template for repair is a very rare occurrence, and it is even more difficult to generate a double knockout, particularly in mammalian cells. Before TALEN editing was available, scientists endured labor intensive screening for heterozygous mutations and then crossed the experimental animals to achieve homozygous knockouts. TALEN technology has proven its versatility in many applications, from single cell models such as yeast to multicellular organisms such as Drosophila, mice, zebrafish, and human stem cells (2, 5).
Other modifications of TALEN
Interestingly, you can pair TALE with other effectors to induce other effects. For example, you can swap FokI nuclease for a Kruppel-associated box transcription repressor (KRAB) for gene-silencing experiments (6). It has been hypothesized that KRAB repression activity might work through mediating negative cofactors (9). On the other hand, if you want to over-express certain genes, you can try to hook up TALE with a synthetic VP64 domain, which is made of the minimal subunits from the Herpes Simplex-1 virus (HSV-1) VP16 domain that recruits endogenous activators of gene expression (7). Even more interestingly, you can add a light switch called cryptochrome2 (CRY2) with a connector to VP64 to make a light-activated gene expression model (3). The way it works is that the specific wavelength of light will change the conformation of the CRY2 and binds to the connector (C1B1). C1B1 will then attract the VP64 domain (3). This is an ideal model for inducing/shutting off temporal expression of your gene of interest, as compared to other gene over-expression models.
Conclusion
There you have it! TALEN is wonderful for molecular genomic engineers and I am sure each day, newer and better ways to utilize TALEN will be revealed. If you are interested in finding out the differences between TALEN and CRISPR are, feel free to check out the following articles at Bitesize Bio:
- CRISPR Technology Explained: Towards a CRISPR genome!
- Getting Started in Genome Editing with CRISPR
- CRISPR Genome-Editing – What You Need to Know to Get Started
References
- Wright et al. TALEN-mediated genome editing: Prospects and perspectives. Biochemical Journal. 2014; 462(1):15-24. DOI: 10.1042/BJ20140295
- Boettcher et al. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol Cell. 2015; 58(4):575–585. DOI: 10.1016/j.molcel.2015.04.028
- Nemudryi et al. TALEN and CRISPR/Cas Genome Editing Systems: Tools of Discovery. Acta Naturae. 2014;6(3):19–40. PMID: 25349712.
- Boch et al. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. 2009; 326(5959): 1509-1512. DOI: 10.1126/science.1178811
- Ding et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell; 2013. 12(2):238–251. DOI:10.1016/j.stem.2012.11.011
- Rauscher et al. Krüppel-associated boxes are potent transcriptional repression domains. PNAS. 1994; 91(10): 4509-4513. DOI: 10.1073/pnas.91.10.4509
- Perez-Pinera et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10(3):239–242. DOI:10.1038/nmeth.2361
- Moscou et al. Xanthomonas bacteria use an amino acid–based code to target effector molecules to specific DNA sequences. 2009; 326(5959): 1501. DOI: 10.1126/science.1178817
- Peng et al. Biochemical Analysis of the Kruppel-associated Box (KRAB) Transcriptional Repression Domain Biol. Chem. 2000; 275(24): 18000-18010. DOI:10.1074/jbc.M001499200
You made it to the end—nice work! If you’re the kind of scientist who likes figuring things out without wasting half a day on trial and error, you’ll love our newsletter. Get 3 quick reads a week, packed with hard-won lab wisdom. Join FREE here.