Plasmid maps are not intuitive to read. And if the first time you see one is during your graduate studies, it can be a bit embarrassing to admit that you don’t understand what you’re looking at. Plus, you run the risk of really messing things up when you try to design and create your plasmid constructs.
Don’t worry. We have a helpful introduction to deciphering plasmid maps.
Familiarizing Yourself with Your Plasmid Map of Interest
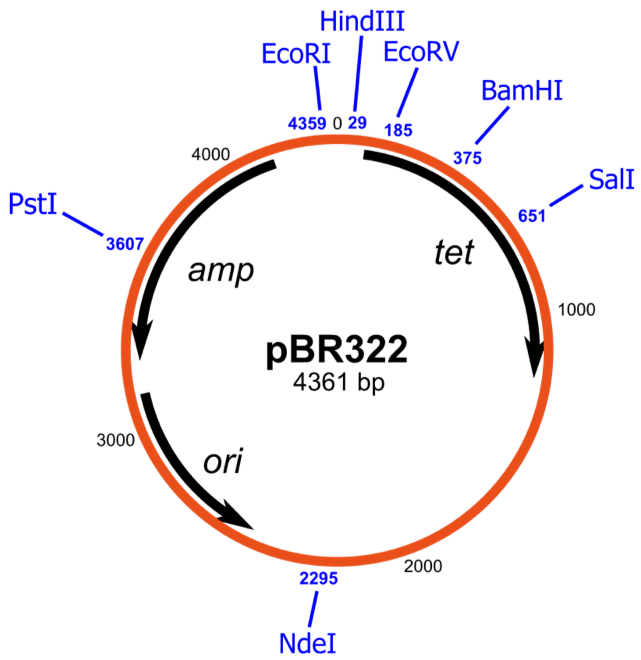
Let’s start with a classic plasmid: pBR322. [1] It is often used as a backbone for derivative vectors because it has all the features needed for successful cloning (Figure 1).
From the map center, we can learn that the size of the linearized plasmid is 4361 base pairs (bp).
The blue text around the outside of the map represents restriction sites. These are the positions where the plasmid will be cut if you incubate it with the corresponding restriction enzyme.
Before you start working with any plasmid, it is advisable to linearize it by cutting with a restriction enzyme and sequencing it via PCR to check that the advertised size and sequence are correct. It is better to sort any plasmid issues out at the start of a project rather than mid-way through.
Where Can I Find the Restriction Sites?
Restriction sites for corresponding enzymes are shown as vertical lines with the position of the starting nucleotides. The sites should be unique, but it pays to check, as derivative vectors often contain additional forgotten sequences.
The black arrows show the direction of transcription, which is essential for cloning. If you clone your gene of interest in the middle of another gene, make sure that both of them are transcribed in the same direction.
Otherwise, the native promoter can interfere with your gene expression.
If you need a refresher on the basics, read this article that explains transcription, translation, and the central dogma of biology.
What Does “Ori” Mean?
Plasmid maps and sequences will always include an ori which is short for the origin of plasmid replication, and if you are looking at a linear plasmid sequence, it will be at the start. Whatever you do, don’t change it! Once a plasmid is unable to replicate, it is useless.
Another thing to remember about the ori is that plasmids with the same origin are often incompatible. This means that you will not be able to maintain two pBR323-derived vectors in one cell even if they have genes for different antibiotic resistances on them.
For example, the pBR322 ori is also in pUC18, among the most common plasmids used in eukaryotic vectors.
Where Can I Find Antibiotic Resistance Genes?
Many plasmids include antibiotic resistance genes, which allow for the selection of colonies expressing the plasmid.
pBR322 has two antibiotic resistance genes: tet (tetracycline resistance) and amp (ampicillin resistance). These genes encode an efflux pump (tetR) and beta-lactamase (ampR) that excrete tetracycline and ampicillin from the cell, respectively. Tet and amp are read in different directions.
Bear in mind that the enzyme beta-lactamase is not specific in detoxifying penicillin-derived antibiotics. So even if you have two plasmids with different origins of replication, you will not be able to select two plasmids at the same time if one expresses a methicillin resistance gene and the other expresses an ampicillin resistance gene.
This is because both plasmids will be subject to the same selection pressure, and the bugs will probably kick one of the plasmids out if it’s unnecessary to their survival.
Where You Cut the Plasmid Matters—A Lot
When using restriction enzyme sites to clone your gene of interest into your plasmid, look carefully at which sites fall within your antibiotic resistance gene. For instance, PvuI cuts in the middle of AmpR, and BamHI cuts in the middle of TetR. And, as we all know, disruption in a gene will lead to inactivation of gene function–in this case, antibiotic resistance.
And you also need to check that your chosen restriction enzymes do not cut your insert. Check if your insert contains a restriction site matching your chosen restriction enzymes.
How Does Replication Start and Stop?
In addition to genes, plasmids often include transcription promoters and terminators derived from E. coli phages. Promoters from phages SP6 and T7 are often used for in vitro RNA amplification. They require phage polymerases and are, therefore, inactive in vivo.
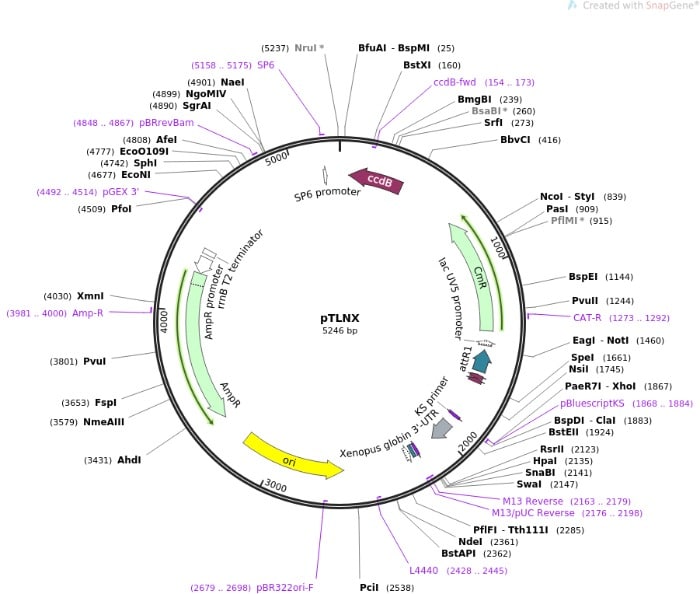
Figure 2 shows the plasmid map of pTLNX, a Xenopus oocyte expression vector. In addition to the familiar pBR322 origin and antibiotic resistance genes AmpR and CmR (chloramphenicol resistance), there are also SP6 and lacUV promoters present.
Downstream (towards the 3′ end) from the SP6 promoter, the rrnBT2 terminator allows efficient termination of genes cloned into the multiple cloning site. [2]
The pTLNX vector also has a gene for plasmid selection (ccdB), the virus SV40 nuclear localization signal, and the Xenopus globin 3′ untranslated region (UTR) that allows for high expression levels of cloned genes.
Know the Source of Your Plasmid
Homemade maps are often unreliable as they omit features that may be critical for your experiment. If you need a vector map, it is better to use known map repositories such as Addgene and websites of companies that sell your vector of interest.
Coming Full Circle: Plasmid Maps Summarized
And that was plasmid maps for the uninitiated. You know where to look for restriction sites, the origin of replication, plasmid size, antibiotic resistance, transcription direction, and more!
And remember, homebrew plasmid maps are always evolving, so it is likely that your contributions will be left for your future colleagues. Please include as many details as possible in your map! Happy map reading and drawing.
Originally published March 2019. Revised and updated July 2023.
References
- Balbás P, Soberón X, Merino E, Zurita M, Lomeli H, Valle F, Flores N, and Bolivar F (1986) Plasmid vector pBR322 and its special-purpose derivatives — a review. Gene 50:3–40
- Geertsma ER and Dutzler R (2011) A versatile and efficient high-throughput cloning tool for structural biology. Biochem 50(15):3272–8