Generating a DNA vector library of single and/or compound mutants for a target protein can be a daunting task. If you’re lucky and work in a well-funded lab, you might outsource this process via gene synthesis. Most of us though, need to do it the old-fashioned way.
Traditional QuikChange™
Traditionally, there are many steps you need to complete for Quickchange™. You need to:
- Order a series of DNA primers to generate mutation(s)
- Set up QuikChange™ PCR,
- Transform competent cells
- Stab colonies for mini overnight cultures
- Purify resulting vector DNA
- Submit samples for sequencing
This process is time consuming in itself, taking up to a week to determine if you achieved the desired mutation. Moreover, due to the likelihood of failed incorporation of mutations, the actual costs can add up from primer optimization and repeated sample sequencing. Not to mention the mounting frustrations over waiting to get your desired vector in order to test the expression and purification of your mutant protein.
Eureka
I recently embarked on a similar endeavor, in which I needed to test a series of single and double mutants covering three residues of interest. My PI came up with a brilliant plan to expedite generating these mutations. For this method you need:
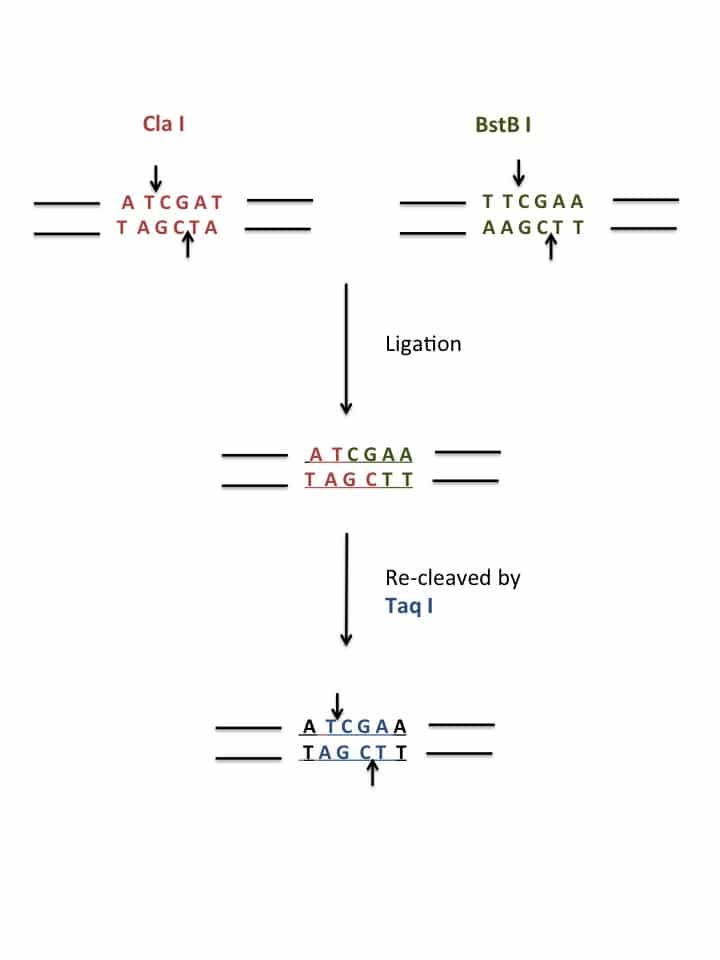
- Unique restriction sites within your gene that isolate the region of interest
- Small gene synthesis products containing your mutations
How to do it
First
Map all unique restriction sites within your vector and identify two sites that, when treated with endonucleases, excise a small fragment (100-200 bp) containing the region of interest to mutate.
Second
Design a 500 bp gene synthesis product (following your chosen company’s recommendations), that contains the same target sequence from above, but including your desired mutation(s) flanked with the same restriction sites (I managed three mutant sequences within the 500 bp limit). Include a generic promoter and terminator (T7, M13, SP6..) so that you can PCR amplify the full-length product to have ample substrate to generate digested inserts.
Third
Set up a PCR reaction to amplify the gene synthesis product. Digest the PCR product and vector + gene of interest with your chosen restriction endonucleases. Gel purify the linearized vector and column purify the digested insert (the insert is small, and therefore will be difficult to visualize on an agarose gel). Your gene synthesis product is now a heterogeneous mix of insert fragments containing different mutant sequences of interest.
Note: if you design multiple gene synthesis products, treat each 500bp substrate individually.
Fourth
Set up ligations, transform, pick colonies (twice as many colonies as fragments were ligated), mini-prep, and sequence to identify the different variants.
Limitations
One of the biggest limitations to this technique involves luck. Not every gene will have convenient restriction sites that you can use to isolate your region of interest. On the contrary, your gene may have multiple cut sites for one or more restriction enzymes, and therefore will prohibit targeting a specific region of interest.
Not all restriction enzymes efficiently cut on short DNA sequences, therefore it’s possible that not all desired inserts can be achieved. Moreover, this technique relies on both restriction enzymes having equal activity in the same buffer (NEB#2, CutSmart…). If your restriction enzymes require different buffers, this approach may not be feasible. Column purifying the intermediate digested gene synthesis fragments after each enzyme is a possible solution, however this has not been tested.
I successfully cloned three mutants with this technique. I first sequenced six colonies, which resulted with identification of two of the three target mutants. I sequenced three more colonies and identified the third target mutant. The value in this approach is that DNA sequencing provides results overnight. When I realized I needed to identify the third mutant, I was only two days removed from this result. If I were relying on QuikChange, it would have taken another week to determine if the mutation was incorporated.