Want to measure how much mRNA you have in a particular sample? Easy! Make some cDNA, add some fluorescent DNA-intercalating dye, pop it into a quantitative real-time PCR (qRT-PCR) machine and Bob’s your uncle! You have your result! Easy right…? Not so fast.
As with any scientific assay, qRT-PCR requires some optimization. First, you need to test your primers for specificity and efficiency and second, and most commonly overlooked, you need to optimize and select your housekeeping gene.
Why Do I Need a Housekeeping Gene?
Unless you are assessing absolute quantification by generating a standard curve, you generally are assessing relative expression (the difference between two samples). To be able to compare the expression level, you need to ensure that their initial input was the same, because qRT-PCR will exponentially increase your desired product. Owing to the sensitivity of qRT-PCR, slight differences in the DNA content of samples caused by pipetting errors, inefficient cDNA synthesis and inaccurate quantification will lead to a quantifiable difference. Thus, a good housekeeping gene provides a point of reference to normalize all values to and ensure that the effect of varying DNA content is minimized.
What is a Good Housekeeping Gene?
The definition of a good housekeeping gene, is one that has stable expression in varying conditions, thus can be used to normalize other influences which may skew the result. Common housekeeping genes such as beta-actin, HPRT, GAPDH and 18S have all been shown to have unstable expression under different conditions and disease states, suggesting that they may not be suitable for use in all situations. In fact, none of these genes were present in the top 50 most stably expressed genes in one study! To confidently be able to ensure consistent and correct results, you need a housekeeping gene whose expression is stable across all of the parameters you are assessing. Keep in mind; this will likely be different across different cell lines too!
How Do I Choose the Right Housekeeping Gene?
Personally, I tested the expression of seven different housekeeping genes in two different cell lines across five different experimental treatments (Figure 1). Although I didn’t see huge deviations from the average Ct value, some values were a full 1 Ct away from the average! This could equate to a twofold expression difference that may cause your data to mislead you. Interestingly, cell line 1 exhibited much higher Ct variability than cell line 2, highlighting that the ‘one size fits all’ approach to housekeeping genes may not be the best. If you have a look at the data, in cell line 1, ACTB (i.e. actin) has one of the highest rates of variability, whereas in cell line 2, it has one of the lowest. From the data, my best housekeeping options for cell line 1 would be GUSB or GAPDH, whereas actin or GAPDH would be more suitable for cell line 2.
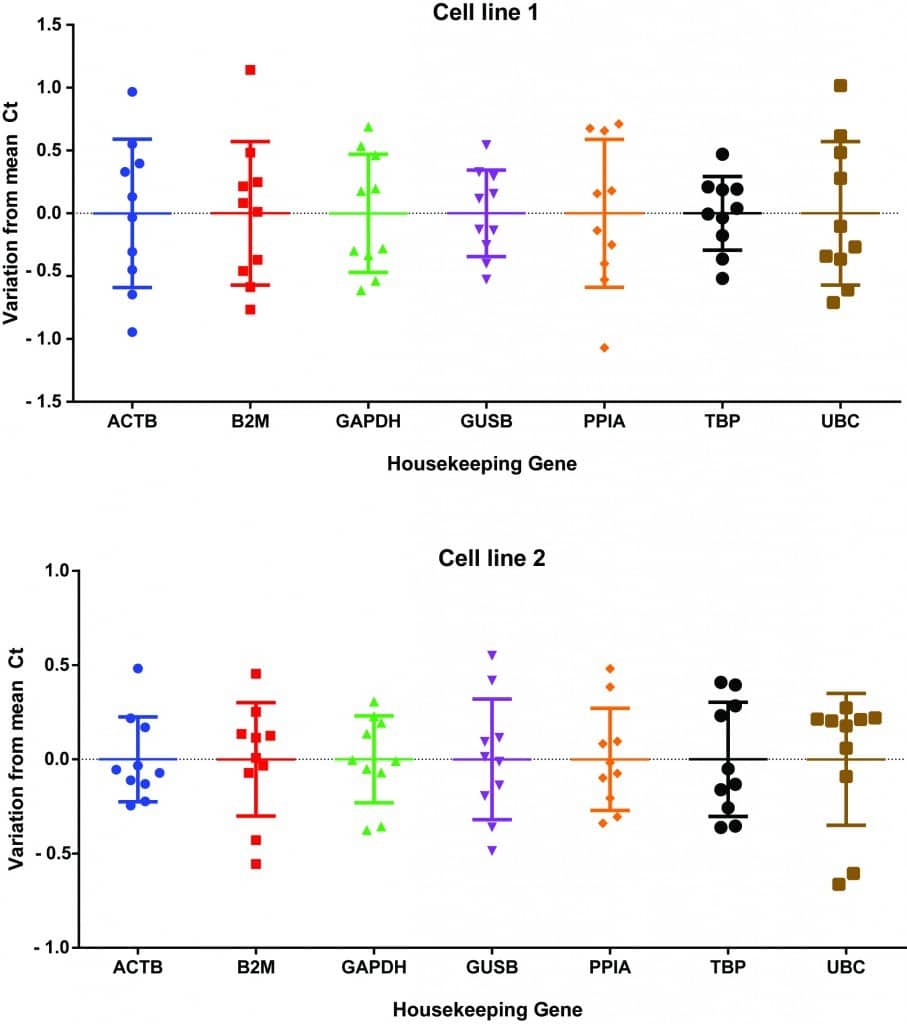
Figure 1. Test of seven different housekeeping genes across two cell lines.
So What Should I Do?
I highly suggest running a panel of housekeeping genes in cell lines and treatments that you use often. There are even PCR arrays you can purchase to help test several housekeeping genes if you are so inclined. There are literally thousands of housekeeping genes to choose from and you can also use the huge amount of gene expression data available online to better select a housekeeping gene. In fact, using more than one housekeeping gene may be the best method.
So remember, although qRT-PCR can be a quick and easy technique to assess relative difference in mRNA transcript expression between samples, picking an inappropriate housekeeping control can drastically skew your data and ultimately lead you down the wrong path. An extra optimization step is all it takes to produce consistent and experimentally sound data – and after all that effort it takes to even get an experiment to work, it’s worth it!
References
Guo et al. ACTB in cancer. Clin Chim Acta 2013;18(417):39–44.
Kang et al. The housekeeping gene hypoxanthine guanine phosphoribosyltransferase (HPRT) regulates multiple developmental and metabolic pathways of murine embryonic stem cell neuronal differentiation. PLoS ONE 2013;8(10):e74967.
Cummings et al. Glyceraldehyde-3-phosphate dehydrogenase is an inappropriate housekeeping gene for normalising gene expression in sepsis. Inflammation 2014;37(5):1889–94.
Craythorn et al. An RNA spiking method demonstrates that 18S rRNA is regulated by progesterone in the mouse uterus. Mol Hum Reprod 2009;15(11):757–61.
de Jonge et al. Evidence Based Selection of Housekeeping Genes. PLoS ONE 2007; 2(9):e898.
Eisenberg & Levanon. Human housekeeping genes, revisited. Trends Genet 2013; 29(10):569–74.
Vandesompele et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3(7).







