The following question was emailed to Bitesize Bio by Beheroze Sattha and I gladly took up the challenge, and I immediately knew the answer. Or so I thought.
After delving extensively into Pubmed, Genes V (I know, I need a new version) and Molecular Cloning I have come up with an answer, but it is not what I originally thought and it turns some aspects of my understanding of plasmids, cloning and transformation on their head. So, although this is my best answer I would love to hear your thoughts on this.
Here is Beheroze’s question:
I was wondering if a single E. coli can take up two plasmids. I am cloning a section of the mitochondrial genome to look for SNPs. After cloning and
sequencing, I notice that some sequences show mixtures, eg Y or R. Does this mean that that particular E.coli had taken up two plasmids, one with a C and the
other with (SNP) T, which when sequenced show up as Y.
My initial reaction was that each E.coli cell can only be transformed by one plasmid molecule, which is then propigated to make a clonal population of bacterial that contains only a single plasmid species. That is the common wisdom as I understand it. But is this true? And if so, how does it happen?
Again, my initial reasoning was that it is extremely rare for a single E.coli to be transformed by two plasmid molecules in a single transformation. But is this true? Well, no it’s not.
Double transformation is not very rare at all!
In your competent cell preparation, only a fraction of the cells are actually competent to take up bacteria and a sub-population of those can readily take up two or more plasmids in a single transformation.
This was shown for CaCl2 transformation of E.coli by Weston et al in 1979 [1] and more recently for electroporation [2]. Weston et al constructed dose response curves (conc of plasmid DNA vs number of transformants obtained), which showed that only 0.26% of the cell population was transformable, and that only 10-20% of those competent cells (so about 0.052%) of the population could take up two or more plasmid molecules. In the more recent work on electroporation, Goldsmith et al also showed that it is possible for cells to be transformed by two independant plasmid molecules in one transformation, and the frequency of double transformations even increases exponentially with the plasmid concentration.
So double transformation – entry of two (or more) plasmid molecules into a single E.coli in a single transformation procedure, is not all that uncommon. In fact according to Weston et al’s statistics, up to 20% of your transformants could be double transformants.
But what about plasmids that carry the same replicon?
But in Beheroze’s experiment, the clones have different inserts on the same plasmid backbone – they all have the same origin of replication. Conventional wisdom says that if two of those plasmids enter a single E.coli during a transformation, plasmid incompatibility dictates that they will not both be propigated.
Plasmid incompatibility is defined as the failure of two plasmids co-resident in the same cell to be stabily inherited. Plasmids with similar origins of replication, and therefore similar systems for regulating the replication and seggregation of the plasmid, are said to be in the same “incompatibility group” and cannot both be passed down consistently to daughter cells. This is because the copy number of a plasmid is tightly regulated by the replicon and having two plasmids with the same replicon interferes with this regulation because they share the regulatory factors required for replication or partitioning during cell division. Effectively, the two plasmid clones compete for the available factors required for replication and partitioning.
How does plasmid incompatibility take effect in a double transformant?
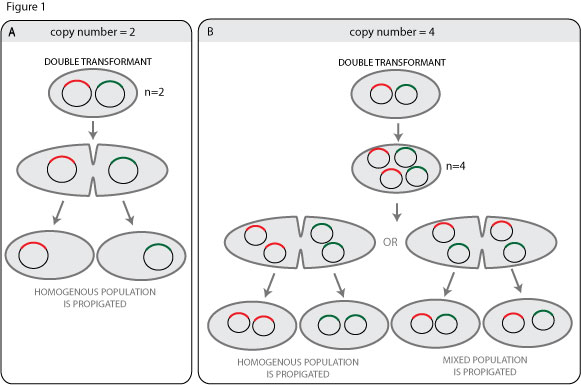
A simple way to think about this us to imagine a plasmid with a copy number of two (see figure 1A). The regulatory machinary of the origin of replication is set to keep the copy number at two, but in a double transformant containing two different plasmid clones with the same origin of replication the copy number of the replicon is already two, so no replication will occur. The two plasmids will then be seggregated to different daughter cells after cell division, so each daughter cell will contain a homogenous plasmid population.
This is all very well, but if the cells are plated before they have a chance to divide, this could –to my mind– result in a clone containing two different plasmid clones. Moreover, most plasmids have a copy number higher than 2, so the situation is more complicated. Even with a copy number of 4 the chances of propigating a heterogenous plasmid population is increased, it would seem.
The commonly understood wisdom is that the two plasmid clones will have small differences that cause one to have a faster replication rate, or increased toxicity, over the other. This is said to cause the plasmids to be replicated assymetrically, contributing to the eventual loss of one of the clones.
This is apparently true in many cases, but a 2007 paper from Velappan et al [3] reports that in double transformants containing two plasmids with identical origins of replication can be stably maintained together in the same E.coli for many days, especially if a high copy number origin (such as pUC) is used.
If you take this is the context of a transformation, to me this seems like it is entirely possible that Beheroze has obtained a colony from that contains a mixed plasmid population. This could have arisen from a single E.coli taking up two different clones during the transformation and being propigated either in the same cells, or in separate populations (or both) as the colony grew.
How can double transformants be avoided?
From this it would seem that in any experiment where a mixed plasmid population is being transformed and a homogenous cloned is desired, double transformations should be borne in mind, diagnosed and avoided where possible.
1. Use low concentrations of plasmid. Goldsmith et al showed that double transformation increased exponentially with plasmid concentration, so keeping the plasmid concentration as low as possible, given the constraints of your experiment, should help.
2. Check clones using colony PCR or sequencing. Colony PCR will work especially well if the PCR will give distinct bands for positive and negative clones.
If you have a double transformant it will give two bands. Beheroze has identified double clones by sequencing the resulting plasmid preps and noticing that
the mixture might contain two different sequences.
3. Choose more than one clone. If you do have some double transformants, they should be in a minority if you choose a bank of positive clones. The singly-transformed positives should be easily indentified by colony PCR or sequencing.
4. If you don’t have a large number of positive clones — re-isolate. Positive clones should be isolated and used to innoculate a fresh culture, plated at low density on an agar plate and individual colonies re-analysed. If the original clone consisted of a mixed population of two cell types containing different plasmids, either due to double transformation or plating at too high a density, this will resolve the two (which is the other possible explanation for Beheroze’s result). Alternatively, you could isolate the plasmid(s) from the positive clone and re-transform. If your plasmid population was mixed, you will get a plate of (mostly) single transformants containing one or other of the originally co-transformed plasmid.
I have to say that this has been a real eye-opener for me, but I like it when I can identify a perception that I hold close without any evidence and do the work to illuminating it. However, I may be missing something here, so if you can provide further illumination, or have any questions, please feel free to leave a comment.
References
1. Weston A, et al Simultaneous transformation of Escherichia coli by Pairs of Compatible and Incompatible Plasmid DNA Molecules. Mol Gen Genetics (1979) 172 p113