This is the first installment in our series on DNA microarrays where I will introduce how this technology is used for the analysis of gene expression, the technology behind this technique, and explain the basics of the technique.
This is to give you a quick immersion if you are new to the subject, and maybe a refresher if you are an old hand at it.
Expressing the Story of Life From the Genetic Code
While humans contain about 25,000 genes, only a fraction of these are actively expressed as mRNAs at any one time. This makes sense since if all the genes were expressed all at once, we would get a garbled stream of unstructured information.
Just like forming words in a sentence, the expression of the genetic code reflects the specific context and the exact message that needs to be stated.
When we talk, we structure our words into a specific string that forms a sentence. Sentences could be arranged into a paragraph, paragraphs into passages, and so on. However, there must be a specific context and purpose to each sentence.
Similarly, specific genes are expressed under corresponding conditions to “update” the functional content of the organism, to respond to internal and external stimuli, resulting in growth, development, and survival. The set of all RNA molecules expressed in a cell at any given time, what it is “saying” at that time, is called the transcriptome.
The Technology
The development of DNA microarray technology in the mid-1990s allowed for the first time to simultaneously profile and study the transcriptome, in other words, to study cells’ real-time “chatter” in more detail. That is, microarrays are a tool for gene expression analysis.
The technology exploited the very same principle that makes nucleic acid so essential to information storage: hybridization to complementary sequences. Hybridization between the cDNA reverse transcribed from a biological sample to a pre-designed complementary DNA probe arranged on a slide, or array, is the basis of DNA microarrays. A microarray, therefore, consists of a pre-designed library of synthetic nucleic acid probes that are immobilized and spatially arrayed on a solid matrix.
The Evolution of DNA Microarrays
Microarrays evolved from a technique known as Southern blotting, where DNA fragments are attached to a substrate and then probed with a known gene sequence.
The first DNA arrays were constructed by immobilizing cDNAs onto filter paper.
However, it was not until 1995 that the first DNA microarrays capable of analyzing thousands of sequences were constructed by “spotting”, or attaching short synthetic probes to designated locations on the solid surface, usually glass or silicon chip (see Figure 1).
How DNA Arrays Are Produced
There are several ways that such spotted arrays can be produced.
Some methods basically use a robot to “print” pre-designed probes that have been attached to fine needles onto a chemical matrix surface using surface engineering (examples include fine-pointed pins, needles, and ink-jet printing).
Other methods employ photo-activated chemistry and masking to synthesize probes one nucleotide at a time on a solid surface in repeated steps to build up probes of specific sequences in designated locations.
Read more about how DNA microarrays are built.
DNA Microarray Measurement of Gene Expression: A Step-By-Step Guide
Let’s walk through a basic protocol for using a DNA microarray for gene expression profiling.
Isolate and Purify
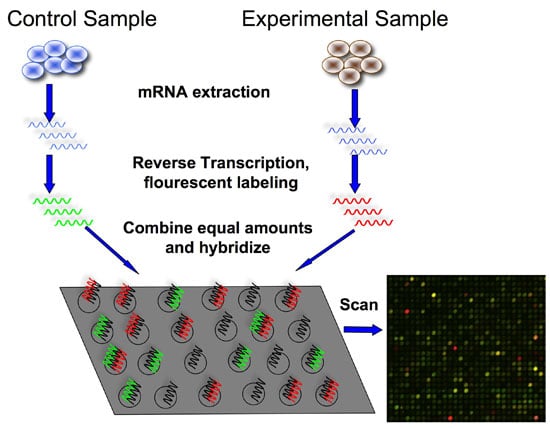
The first step is to isolate and purify mRNA from samples of interest. Since we are interested in comparing gene expression, one sample usually serves as a control, and another sample would be the experiment (e.g., healthy vs. disease)
Reverse Transcription and Labeling
The next step is to reverse transcribe and label the mRNA. In order to detect the transcripts by hybridization, they need to be labeled, and because starting material may be limited, an amplification step is also used.
Labeling usually involves performing a reverse transcription (RT) reaction to produce a complementary DNA strand (cDNA) and incorporating a fluorescent dye that has been linked to a DNA nucleotide, producing a fluorescent cDNA strand.
Disease and healthy samples can be labeled with different dyes and co-hybridized onto the same microarray in the following step. Some protocols do not label the cDNA but use a second step of amplification, where the cDNA from the RT step serves as a template to produce a labeled cRNA strand.
Hybridization
Now it’s time to hybridize the labeled target to the microarray. This step involves placing labeled cDNAs onto a DNA microarray where they will hybridize to their synthetic complementary DNA probes attached to the microarray.
A series of washes are used to remove non-bound sequences.
Scanning and Quantitation
The fluorescent tags on the bound cDNA are excited by a laser and the fluorescently labeled target sequences that bind to a probe generate a signal.
The total fluorescent intensity of the signal depends upon the amount of target sample binding to the probes present on that spot.
Thus, the amount of target sequence bound to each probe correlates to the expression level of various genes expressed in the sample. The signals are detected, the signal intensity is quantified, and used to create a digital image of the array.
DNA Microarrays: An Example Result
If we are trying to calculate relative expression between two samples, each labeled with a different dye (See figure 2, red for the experiment, green for the control), the resulting image is analyzed by calculating the ratio of the two dyes.
If a gene is over-expressed in the experimental sample, then more of that sample cDNA than control cDNA will hybridize to the spot representing that expressed gene. In turn, the spot will fluoresce red with greater intensity than it will fluoresce green. The red-to-green fluorescence ratio thus indicates which gene is up or downregulated in the appropriate sample.
What Have Microarrays Done for Us?
Microarray technology propelled functional genomics, a discipline that strives to identify the role of genes in cellular processes, into the spotlight because it allowed functional analysis of genome-wide differential RNA expression between different samples, states, and cell types to gain insights into molecular mechanisms that regulate cell fate, development, and disease progression.
Microarray data is used for gene expression profiling, which serves as a determinant of protein levels and therefore cellular function between biological samples.
A single experiment can provide information on the expression of thousands of genes, virtually the entire human genome, to compare expression patterns between any two states.
Microarray experiments can indicate which genes are up- or down-regulated between samples from normal and diseased tissue, or two samples in the absence and presence of a certain stimulus.
It is easy to see why this technology might be appealing for understanding complex biological systems as well as drug discovery, disease diagnosis, novel gene identification.
DNA Microarrays Considerations and Caveats
While DNA microarrays have been an excellent genomic tool, and have allowed for some great advances in our understanding of diseases and biological processes, there are some notable limitations to the technology.
Because the targets are preselected, there is little room for discovery (e.g., identification of new gene mutations or unknown sequences), although there are explorative probe design strategies available, which could allow for the detection of mutations.
However, newer techniques, such as RNA-Seq for gene expression profiling, are far more suited to this type of discovery.
Chip redesign can also be costly and time-consuming, meaning this technology can be slow to adapt to newly discovered targets.
Do you still use microarrays in your research? Let us know in the comments below.
Originally published July 13, 2011. Reviewed and updated October 2021.
Further Reading
Trevino V, Falciani F & Barrera-Saldaña H.A. DNA Microarrays: a Powerful Genomic Tool for Biomedical and Clinical Research. Mol Med 13, 527–541 (2007).






