So you’ve isolated your DNA or RNA from your favorite sample. And now, if you are anything like me, the first thing you’ll do is scramble to check the quality and concentration of your extract.
You have a few different options at your disposal to perform this crucial analysis, which will let you know whether you should carry on with your downstream experiments, or give up in dismay and hit the pub.
You know how to do these analyses inside out. But do you know how they work? If you don’t, then you are not alone. But this article, and my next one will walk you through the wonderful world of nucleic acid purity and concentration….
Your main assays for DNA/RNA concentration and purity are…
- Spectrophotometry
- Spectrofluorometry (relax, it’s just measurement for fluoresence)
- Electrophoresis
Each of these methods uses a different approach to interrogate your sample. And you probably use at least one or two of these all the time.
But do you know how they work? And what the relative pros and cons are? Brace yourself, you are about to find out.
Spectrophotometric measurement of DNA and RNA concentration & purity
Spectrophotometry is probably the most commonly used method of them all. It’s the basis of instruments like Thermo Scientific’s Nanodrop and Eppendorf’s Biophotometer. Or, of course, if your boss is very tight with money, you can use a plain old spec.
The basis of the spectrophotometric measurement is the ability of nucleic acids to absorb ultraviolet light. Nucleic acid concentration is measured at 260nm, where they have the highest absorbance.
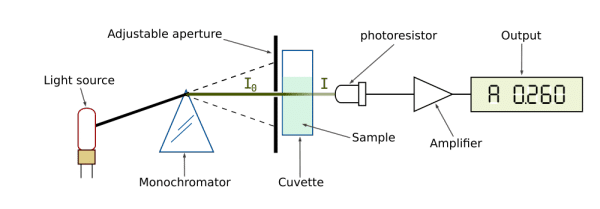
Essentially, a beam of light is directed at the sample with radiant power (I0) and its remaining power after passing through (I1) is measured by a detector.
The ratio I1/I0 is called transmittance (T), which coincidentally reflects the absorbance (A) of the sample, calculated A = log(1/T).
After measuring the absorbance of a sample, the concentration of the absorbing molecule can be inferred using the Beer-Lambert law. Here’s the math bit:
The Beer-Lambert law simply states that absorbance is a product of the length (l) of the path the light takes through the sample (for example, passing through a cuvette), the concentration (c) of the compound in solution and an extinction coefficient (?) that is specific to the molecule in question (also called the molar absorbtivity).
Therefore, the equation A = ? l c can be rearranged to calculate the concentration c = A / ? l.
The extinction coefficient differs between ssDNA (33), dsDNA (50) and RNA (40), which is why you have to supply this information about your sample to the machine.
Spectrophotometers also allow you to measure purity along with concentration. DNA purity is evaluated by the ratio of absorbance at 260nm to 280nm. High quality DNA should have an A260/A280 ratio of 1.7 to 2.0. Other possible contaminants are salt or phenol, which are measured at 230nm. The A260/A230 ratio should be greater than 1.5.
So with one sample, you can measure the absorbance at 230, 260 and 280nm to determine both concentration and purity of your nucleic acids. Be careful though, contamination with different nucleic acids (for example residual RNA, primers or dNTPs) will affect the concentration measurement because they also absorb at the same wavelength.
Ok, that’s enough for today. Go and unfrazzle your brain and come back next week, when I’ll go through spectroflurometric and electrophoretic analysis of your nucleic acid samples.