A comprehensive map of drug targets established that 75% of 1,578 US FDA-approved drugs target human proteins [1].
Proteomics, the comprehensive profiling of proteins within tissues and cells, is critical for understanding the biological function of proteins, their involvement in disease mechanisms, and their potential therapeutic candidacy.
By analyzing the proteomes of relevant cell cultures or clinical samples like formalin-fixed paraffin-embedded (FFPE) and fresh frozen (FF) tissues, researchers can identify biomarkers, monitor treatment responses, and discover therapeutic targets.
In this article, we demonstrate how integrating Adaptive Focused Acoustics® (AFA®) technology into your proteomics workflows overcomes major bottlenecks.
We also explore the benefits AFA provides, including increasing protein yield and coverage, increasing data reproducibility, and achieving automation across diverse sample types.
What is AFA, and How Does it Work?
AFA technology uses high-frequency acoustic energy precisely targeted to a localized zone within the sample vessel, optimizing energy efficiency and minimizing heat generation.
This focused, non-contact energy enables highly controlled cell lysis, allowing for the standardized release of biomolecules, which is crucial for reproducibility and scalability in proteomic workflows.
Major Sample Preparation Bottlenecks in Proteomics
Traditional protein extraction methods, including mechanical and chemical extractions, often suffer from too much variability and a lack of scalability, leading to an expensive bottleneck that limits progress in proteomics.
Obtaining reliable proteomic data is fraught with challenges, particularly during sample preparation. Key difficulties include:
Loss of Material
Sample handling, especially multiple transfer steps, can lead to significant protein loss, especially when starting with small sample volumes. Many proteins of interest are present at low concentrations and are difficult to detect. Sample loss increases the probability of low-abundance proteins falling below the limit of detection.
Time-consuming Processes
Many traditional sample processing methods include time-consuming steps such as overnight trypsin digestion or inefficient deparaffinization of FFPE samples.
Low Yield and Quality
Proteins are sensitive to environmental conditions, and even minor changes in temperature or pH can alter protein quality and yield.
Variation Between Sample Specimens
The massively broad range of biological samples, from cultured cells to tissue biopsies, naturally leads to a variation in protein extraction protocols, limiting compatibility with automation. As a result, performing proteomics on diverse sample types and volumes can become a time-consuming and labor-intensive process for any single lab.
Poor Reproducibility
Achieving consistent results across different runs, instruments, or labs can be challenging due to the factors described above.
These challenges necessitate a more efficient approach to protein extraction, such as AFA, that mitigates the reproducibility and scalability bottleneck.
Examples of How AFA Overcomes Sample Preparation Bottlenecks in Proteomics
Adaptive Focused Acoustics (AFA) technology by Covaris addresses these challenges by using ultrasonic energy in a controlled, non-contact environment.
It reduces the time required for sample preparation and enhances the reproducibility of protein yields across various sample types.
The following case studies demonstrate how major research organizations have integrated Covaris’ AFA technology to increase efficiency and radically improve the output of their proteomics studies.
Overcoming Low Cell Numbers
Traditional, multi-step sample preparation methods are particularly difficult for samples that contain low cell numbers due to the likelihood of losing low-abundance proteins in the process.
To overcome these challenges, researchers at the Mayo Clinic used AFA to extract proteins from HuCC-T1 cells with the Covaris R230 Focused-ultrasonicator [2].
The group compared the AFA-assisted method to the Single-pot, Solid-phase-enhanced Sample preparation (SP3) bead-based digestion protocol, a commonly used method for low numbers of cells due to its capability to minimize sample loss.
The research group showed that the AFA-assisted approach was able to handle wide variations in cell numbers, from as few as 1,000 cells to over 500,000, without loss of efficiency.
The turnaround time from cell lysis to analysis on the mass spectrometer was also much quicker using the AFA-assisted approach. This is due primarily to the accelerated trypsin digestion using AFA compared to overnight digestion for the SP3 method.
Generating High Protein Yields from FFPE Samples
Many research groups use FFPE blocks because they are a cost-effective method for long-term storage of primary clinical samples at room temperature.
Sample preparation from these blocks is challenging because protein yields and sample stability are often difficult to reproduce due to the tissue being fixed in formalin and embedded in wax.
This can introduce multiple crosslinking artifacts that require an aggressive decrosslinking method to eliminate.
The Broad Institute of MIT and Harvard frequently analyzes hundreds of samples from FFPE blocks. To improve their high-throughput sample preparation workflow, the group trialed the Covaris deparaffinization workflow [3].
This method eliminates the need for multiple ethanol and water wash steps and drying of the tissue before adding lysis buffer, saving time by reducing the number of steps.
The group was able to shorten sample preparation time from seven days to just two. Instead of using xylene to dewax the scroll, the Covaris method involves:
- One hour of ultrasonication (for a full plate)
- Decrosslinking with a thermal cycler at 90 °C for 90 minutes
- Another round of ultrasonication for 1 hour (for a full plate) to homogenize the tissue
The AFA setup specifically addressed the challenges of cross-linking and protein accessibility.
Furthermore, by adjusting the duty factor (50% sonication time) and using higher buffer volumes (150 µL), the researchers doubled the yield of protein.
Plus, the group demonstrated that the Covaris methodology is compatible with a wide range of proteomics workflows and instruments, including data-independent acquisition (DIA) and tandem mass tags (TMT) approaches (Figure 1).
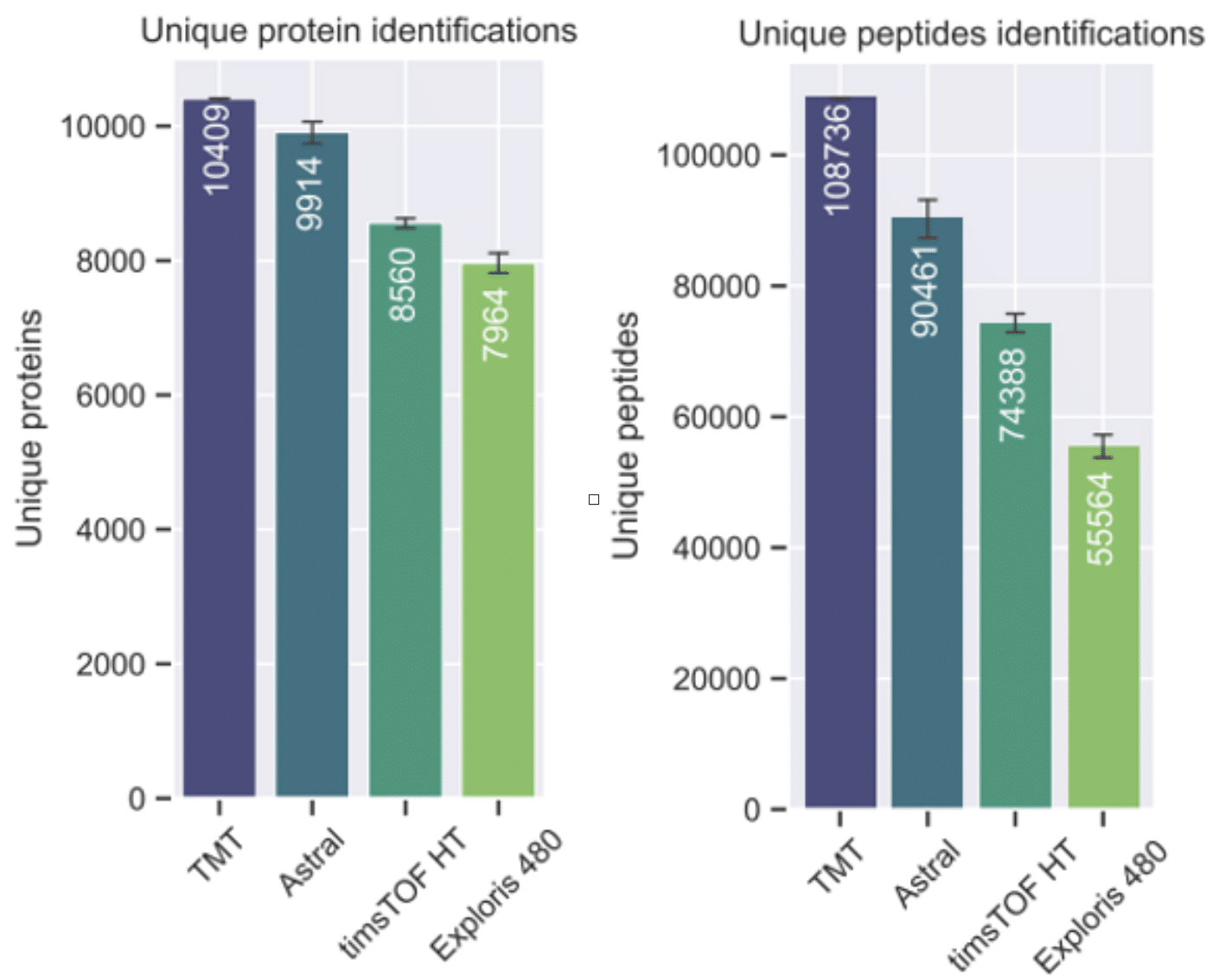
Figure 1. Benchmarking three DIA platforms against TMT. Left: Protein identifications by each method. Right: Peptide identifications by each method [3].
Getting Reproducible Coverage from Multiple Cells and Different Tissue Types
Covaris partnered with Precision Biomarker Laboratories to develop a high-throughput, versatile, and reproducible sample preparation method [4].
Traditional methods that proved unsuitable for automation and high throughput included physical lysis and protein precipitation with organic solvents or strong acids.
The latter in particular presented additional problems with pellet solubilization, automation, throughput, and time-consuming incubation periods.
Part 1: Improving Protein Extraction Efficiency
The first part of developing the AFA-assisted workflow analyzed protein extraction efficiency from HEK293 cells. This part involved two approaches:
- Four quantities of cells (250k, 500k, 750k, and 1 million) were tested to evaluate whether protein yield differed with cell amount
- Extractions were repeated over three days to see if protein yield was reproducible between experiments
HEK293 cells were added to Covaris 8 AFA-TUBE TPX Strips with SDS lysis buffer to a final concentration of 2% SDS and lysed in the Covaris LE220-plus Focused-ultrasonicator.
Protein lysates were digested using automated SP3 and analyzed via LC-MS/MS.
The protein yield increased in a linear trend with increasing cell numbers. Approximately 25 µg protein was isolated from 250k cells, and over 100 µg from 1 million cells.
This yield of approximately 1 µg per 10k cells remained consistent over three days of experimentation, showing significant reproducibility.
Part 2: Assessing Sample Preparation Reproducibility
The second part of this study examined the reproducibility of sample preparation from tissue using a Covaris protocol and was again tested in two ways:
- To see if there was a consistent yield between mouse tissue types, four organs were used: heart, intestine, liver, and lung
- To see if there was consistency in the number of protein groups identified, experiments were replicated over three days
Each tissue type was placed into Covaris Tissue Lysis Buffer (TLB) and processed in the Covaris LE220-plus instrument.
Tissue lysates were digested using SP3, and LC-MS/MS analysis was conducted on a Thermo Scientific™ Orbitrap™ Astral™ using DIA methodology. The number of protein groups identified was consistent across three days in all four tissue types (Figure 2).
The number of protein groups ranged from over 6,000 for the heart tissue to approximately 10,000 for the intestinal and lung tissue.
Further demonstrating the significant reproducibility of samples, the variation between replicates was consistently low over the three days tested, with most of the identified proteome having coefficient of variation (CV) values of less than 20%.
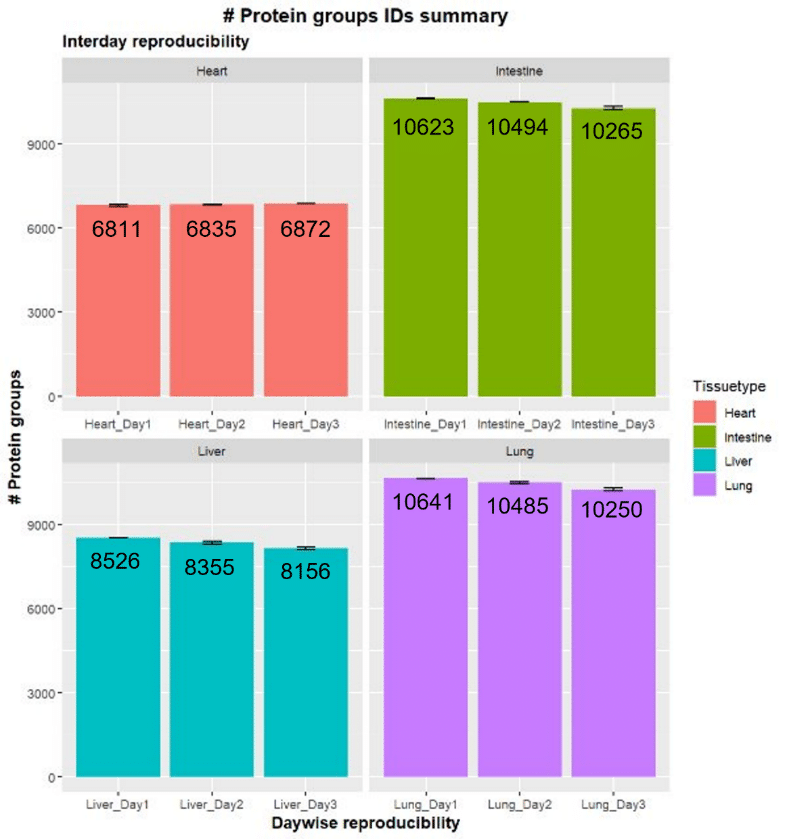
Figure 2. Reproducibility of the number of protein groups identified by LC-MS/MS of four mouse tissue types. Experiments were repeated on three separate days [4].
Achieving Robust, Plate-based AFA for Consistent High-throughput Proteomics
Providing targeted workflows across highly varied sample types and protein input levels is a central proponent of IQ Proteomics’ (IQP) services, so the research team wanted to streamline their processes [5].
Traditional protein preparation techniques have proved insufficient for IQP’s needs because
- They don’t apply to the broad range of samples used
- Their throughput is too low
- They are too slow
The goals of the project were to develop a flexible workflow that could:
- Overcome the sample viscosity associated with cell lysis
- Achieve high recovery from low input amounts
- Do this in 96-well plates to enable integration with automation
Samples were processed in a rack of 96 microTUBE-130 Bead Snap Cap tubes, which are compatible with a wide range of cell numbers. A Covaris R230 Focused-ultrasonicator was situated adjacent to the deck of the Tecan Fluent® Liquid Handler (Figure 3).

Figure 3. Photo of the R230 and the Tecan setup at the IQP laboratory.
Covaris Tissue Lysis Buffer (TLB) was added to the tubes in the rack, which was placed in the Covaris R230 for lysis.
The rack was then moved automatically by a robot to a UV/VIS Spectrometer for a BCA protein quantification. Cytiva Sera-Mag™ beads and ethanol were added for precipitation prior to the SP3 digestion workflow.
By automating the SP3 workflow on the integrated Covaris and Tecan instruments, the research team was able to identify a higher number of peptides over a wide range of sample amounts.
More peptides were identified using the integrated Covaris method than using methanol-chloroform precipitation (for a 20 µg starting sample mass) (Figure 4).
Plus, the integrated Covaris method delivered higher reproducibility across replicates compared to the methanol-chloroform workflow.
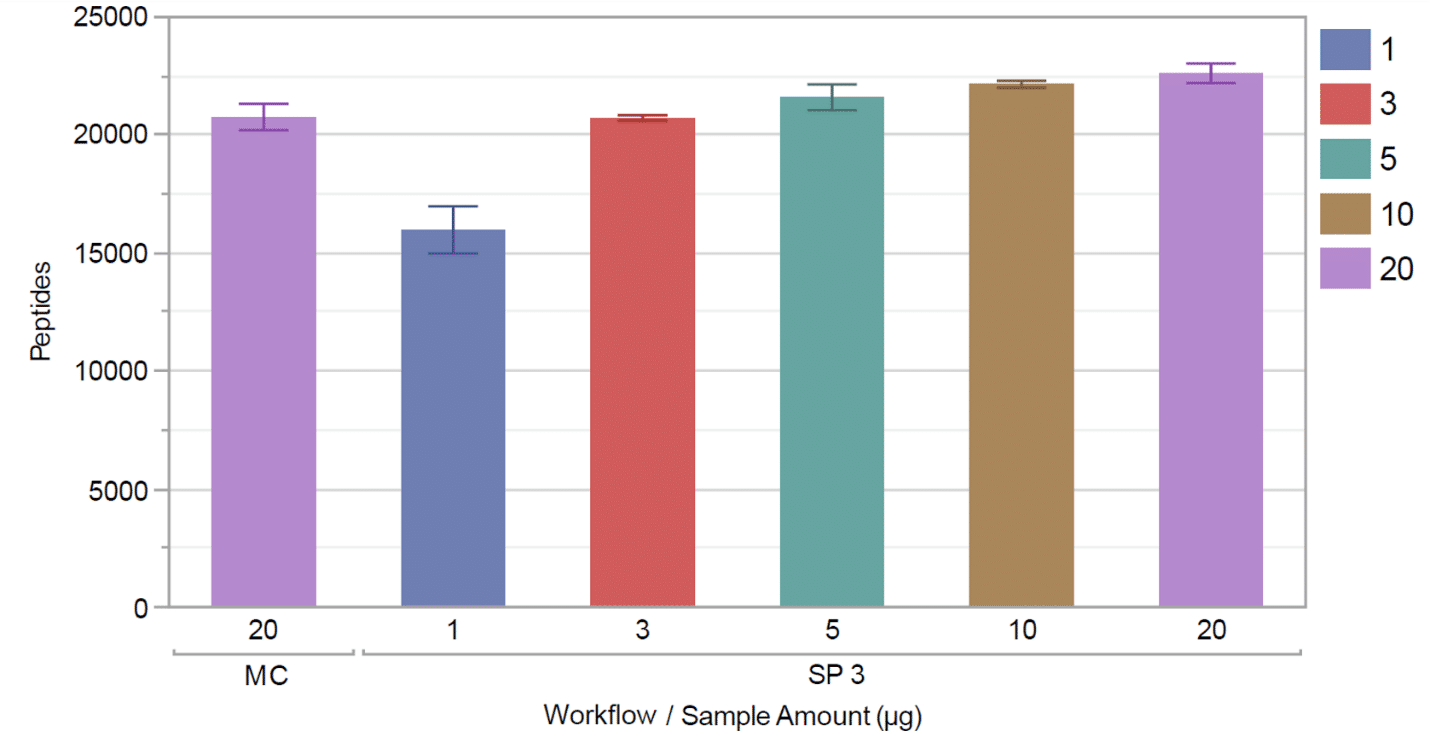
Figure 4. Comparison of methanol chloroform (MC) precipitation with Covaris/Tecan automated SP3 over a range of protein input amounts.
Scaling-up Proteomics Workflows with AFA Summarized
AFA technology greatly assists protein extraction for proteomics by addressing the common issues with sample preparation, such as low protein yields from small sample volumes, low reproducibility, time-consuming workflows, and the challenge of extracting proteins from complex samples like FFPE tissues.
Its ability to provide rapid, consistent, and scalable results provides important benefits to your proteomic studies.
For instance, it can drastically reduce processing time from days to hours and maintain high protein integrity across a wide range of cell counts.
Furthermore, AFA can enhance protein yield and quality, ensuring excellent proteome coverage.
This makes AFA technology indispensable for proteomics, as it ensures the consistent, comprehensive analysis essential for identifying key indicators of disease presence, progression, and response to treatment.
References
- Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI and Overington JP. (2017). A comprehensive map of molecular drug targets. Nature Rev Drug Discov. 16(1):19–34
- Jain A, Charlesworth CM, Sachdeva G, Vasantgadkar S, Bhattacharyya D, Pandey A. (2024, June 2-6). Proteomics Analysis of a Limited Number of Cells by a Rapid and Efficient Workflow using Adaptive Focused Acoustics Technology [Conference presentation and poster]. American Society for Mass Spectrometry 72nd Conference on Mass Spectrometry and Allied Topics (ASMS) 2024, Anaheim, California, United States
- Haines M, Thorup J, Gohsman S, Heil L, Vasantgadkar S, Abarzua L, Newton C, Rohrer D, Hostetter G, Mani DR, Gillette MA, Carr SA, Satpathy S (2024, June 2-6). High-throughput, streamlined processing workflow of formalin-fixed paraffin-embedded (FFPE) tissue yielding up to 10,000 proteins per sample [Conference presentation and poster]. American Society for Mass Spectrometry 72nd Conference on Mass Spectrometry and Allied Topics (ASMS) 2024, Anaheim, California, United States
- Ortiz J, Keoseyan A, Bhosale DS, Hendricks N, Seyedmohammad S, Stotland A, Vasantgadkar S, Abarzua L, Moradian A, Mockus S, Van Eyk J. (2024, June 2-6). High-throughput and automated cellular and tissue lysis using COVARIS acoustic technology for proteomics [Conference presentation and poster]. American Society for Mass Spectrometry 72nd Conference on Mass Spectrometry and Allied Topics (ASMS) 2024, Anaheim, California, United States
- Braun C, (2024, June 2-6). Leveraging Plate-Based AFA For Robust and Consistent High Throughput Proteomics [Conference presentation]. American Society for Mass Spectrometry 72nd Conference on Mass Spectrometry and Allied Topics (ASMS) 2024, Anaheim, California, United States