DNA sequencing is a fundamental technique in modern molecular biology that has revolutionized the study of genes, their functions, and their interactions.
It has helped to uncover the genetic basis of diseases, discover new drugs and therapies, monitor people’s response to these treatments, and shed light on the evolution of life on Earth.
The Maxam–Gilbert sequencing method was widely used in the early days of DNA sequencing. But newer and more efficient sequencing technologies have since taken over.
Yet, it still has some niche uses. Plus, in the historical context of DNA sequencing, it’s hugely important.
So let’s learn how Maxam–Gilbert sequencing works, why it’s less popular than it was, and its present-day applications.
A Very Short History of Two DNA Sequencing Methods
First, a bit of history. In the mid-1970s, two methods were developed to sequence DNA directly. These were:
- The Maxam–Gilbert sequencing method
- The Sanger chain-termination method
In 1980, both Walter Gilbert and Frederick Sanger were awarded The Nobel Prize in Chemistry for contributing to the determination of base sequences in nucleic acids. Technically, each got a quarter of the award, because Paul Berg received the other half for his research into nucleic acid biochemistry, particularly regarding recombinant DNA.
Therefore, Maxam–Gilbert sequencing and the Sanger method comprise the first generation of DNA sequencing methods.
However, while Sanger sequencing is still widespread, Maxam–Gilbert sequencing has been largely forgotten. You might be surprised to know, therefore, that when both methods were discovered, Maxam–Gilbert was the more popular.
This popularity was because scientists could use purified DNA directly, while the initial Sanger method required DNA cloning for the start of each read. Readers will appreciate that cloning can be problematic today, let alone several decades ago when the idea and underlying technology were still new.
How Does Maxam–Gilbert Sequencing Work?
Maxam–Gilbert sequencing is also known as chemical sequencing because chemical reactions, rather than DNA and RNA amplification, are the basis of the method.
It’s quite easy to understand, though, and proceeds in just 4 steps:
1. Preparation of Your Sample
The DNA used in Maxam-Gilbert sequencing is first denatured into single-stranded chains and radiolabeled on the 5′ end, usually with 32P.
2. Cleavage
The next step cleaves the DNA. And this is where the Maxam–Gilbert sequencing gets really interesting.
By taking advantage of piperidine and two chemicals that selectively attack purines and pyrimidines (dimethyl sulfate and hydrazine, respectively), the DNA is cleaved at specific points.
To be more accurate, using different combinations of these chemicals, you can cleave a DNA sequence wherever there is a C, wherever there is a C or a T; wherever there is a G, or wherever there is a G or an A.
So, if you put your sample into these 4 different reaction tubes, you obtain different fragments depending on the combination of chemicals!
3. Electrophoresis and Autoradiography
These reactions are then loaded onto a high-percentage agarose gel to differentiate fragment sizes. The fragments are visualized via the radioactive tag.
4. Reading the Sequence
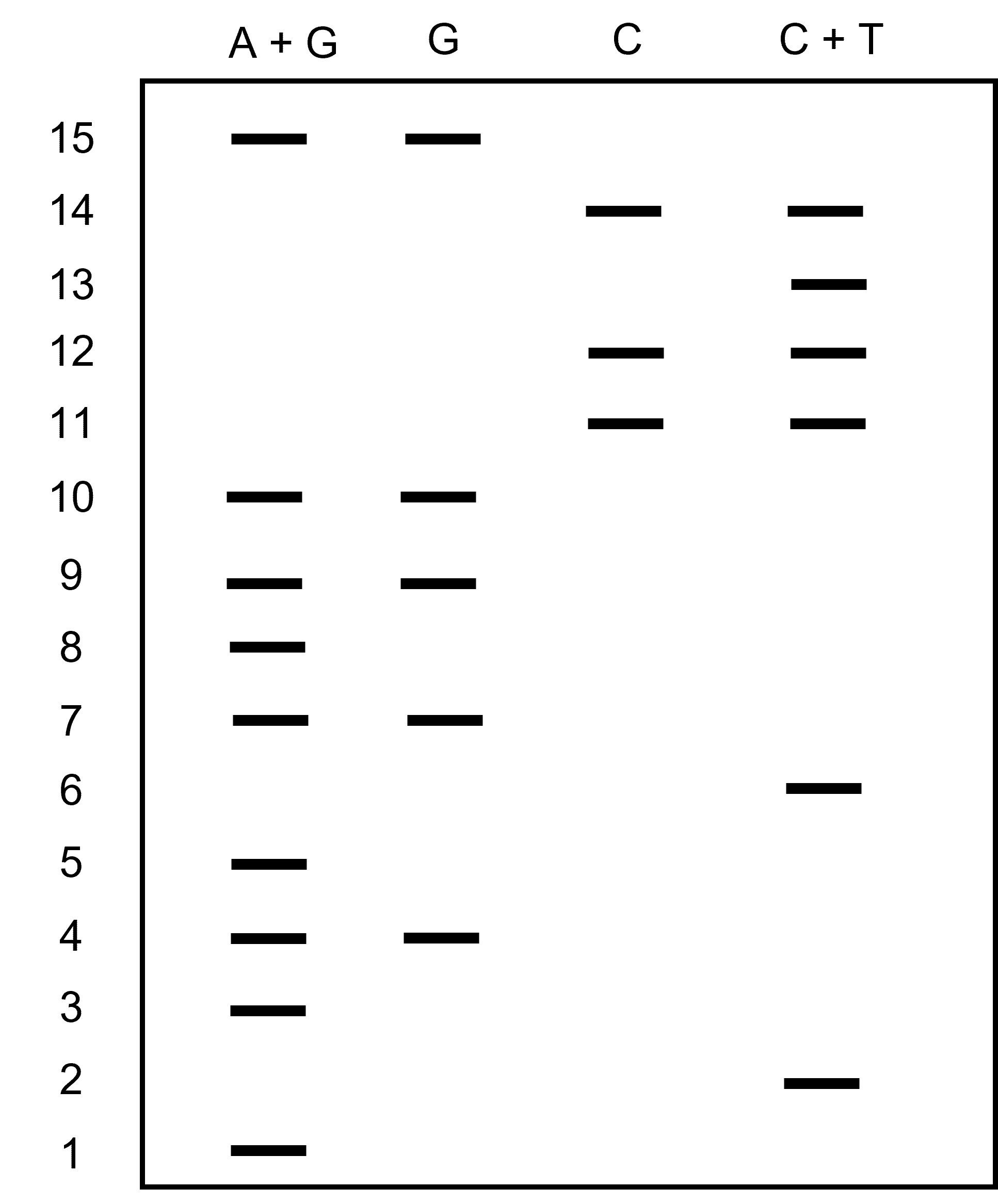
To read the sequence, you begin with the smaller fragments at the bottom of the gel. “Calling” each base involves interpreting the band pattern relative to the four chemical reactions.
For example, if a band in the DNA sequence appears in both the G-reaction and the G+A-reaction lanes, then that nucleotide is a G.
If a band in the DNA sequence appears only in the G+A-reaction lane, then it is an A. The same decision process works for the C-reaction and the C+T-reaction lanes. Sequences are confirmed by running replicate reactions on the same gel and comparing the autoradiographic patterns between replicates.
Then the process repeats like you are solving a puzzle.
Figure 1 shows an example gel. Can you ‘read’ the gel?
Why Did It Lose Popularity?
Maxam–Gilbert sequencing, although based on simple principles, came with a whole lot of trouble. Here are a few of its major drawbacks.
It’s Slow
First, it was time-consuming. And that was supposing that everything went well on the first try. A lot of steps in the method could cause problems. Examples of potentially problematic steps include:
- Radiolabeling.
- The cleavage reactions.
- Preparing the gel.
- Electrophoresis.
- Developing the X-ray film to visualize results.
Plus, you can’t easily parallelize the method. So, you can only confirm about 200–300 bases of DNA every few days!
It Uses Dangerous Chemicals
Working with radioactive isotopes and hazardous chemicals is never ideal. Maxam–Gilbert sequencing uses both.
In particular, 32P is a nasty isotope as they go. It’s a high-energy beta emitter with a short half-life. Or in English, it can put a lot of energy into your tissue and do so quickly.
Its molar activity is 338.61 TBq/mmol (338610000000000 disintegrations per second per millimole), and its half-life is about 14 days.
And hydrazine is a potent neurotoxin that can cause organ damage. The less of that used, the better!
It’s Comparatively Information-Poor
Next-generation sequencing methods, such as the Illumina dye method, offer genome-wide sequencing data. And Sanger sequencing is the incumbent method for plasmid sequencing—a fundamental service required by almost all biology research labs.
Partly because of the drawbacks mentioned above, and partly because of the intrinsic limitations of the technique, Maxam–Gilbert sequencing cannot compete with these methods.
What Is Maxam–Gilbert Sequencing Used for Today?
While the Maxam-Gilbert method is not used as much as it once was, it is still used in some specialized applications where aspects of the process (namely the chemical cleavage step) make it useful
DNA Footprinting
DNA footprinting enables the Identification of sequences of DNA involved in protein-DNA interactions, and Maxam-Gilbert sequencing can be used to do the sequencing step. After labeling the putative protein-binding DNA sections, they are incubated with the protein in question and cleaved as per the Maxam–Gilbert protocol.
The protein will protect the DNA to which it is bound from cleavage while unbound fragments get chopped up.
The DNA is then unbound from the protein and then ran on a gel along with cleaved control fragments. Comparing the pattern of DNA fragments with and without the protein-bound, you can figure out the sequence of DNA that the protein binds to. [1]
Identifying DNA Modifications
DNA can get modified, which can be significant in certain contexts (epigenetics, anyone?). Example modifications include methylation and acetylation.
Again exploiting the cleavage steps, you can cleave the DNA fragments at positions you suspect to be modified, sequence them, and compare these to a series of control fragments. This enables you to figure out if your DNA fragment has been modified.
Structural DNA Analysis
Similarly, enough is known about DNA structure in relation to certain short sequences of base pairs that you can use Maxam–Gilbert sequencing to obtain such granular sequence information that you can begin to make structural inferences about the DNA. Not bad, eh?
Usefulness in Obscurity: Summing It Up
And that’s a summary of Maxam–Gilbert sequencing. How it works, the newer and more powerful sequencing methods like next-generation sequencing that have consigned to relative obscurity, and some of its remaining applications.
The somewhat tedious and hands-on chemical steps involved make it ideal for detecting modifications to and, indeed, the sequence of small fragments of DNA. This, in turn, is useful for characterizing DNA-binding proteins.
Down but not out, as they say. And without a doubt, the grandfather of Genomic sequencing methods.
Have you learned something interesting from this article? Still use Maxam–Gilbert sequencing in your research? Let us know in the comments section below.
Originally published July 2017. Revised and updated March 2023.
Reference
- Sidote DJ et al. (2008) Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure 16:727–35