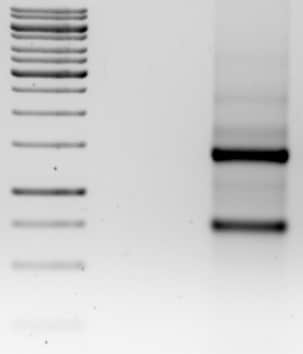
As a follow-up to our article about ethanol precipitation of DNA and RNA, this article explains the differences between DNA precipitation in ethanol and isopropanol, helping you to figure out which method is the best choice for your experiment.
Requirements for DNA Precipitation
First, let’s review the components we need to precipitate DNA or RNA with ethanol:
1. Salt to neutralize the charge on the nucleic acid backbone. This causes the DNA to become less hydrophilic and precipitate out of solution.
2. Ice to chill the sample. Lower temperatures promote the flocculation of the nucleic acids so they form larger complexes that pellet under the centrifugal forces of a microcentrifuge.
3. A nucleic acid concentration high enough to force the DNA out of solution (if the concentration is not high enough, you can add a carrier nucleic acid or glycogen to enhance the recovery).
4. A microcentrifuge to pellet the sample.
Isopropanol Vs. Ethanol: DNA Solubility
DNA is less soluble in isopropanol so it precipitates faster even at low concentrations. [1] The downside however is that salt will also precipitate in isopropanol. With ethanol, the DNA needs to be at a higher concentration to flocculate but the salt tends to stay soluble, even at colder temperatures.
DNA precipitates in 35% isopropanol and 0.5 M salt. Using ethanol, the final concentration needs to be around 75% with 0.5 M salt. So for the typical precipitation protocol, isopropanol is added from between 0.7–1 volumes of sample, and ethanol is added at 2-2.5 volumes of sample.
Choosing the Right Solvent: Sample Volume
If you are precipitating small volumes of DNA, and you can fit the required amount of solvent into the sample tube, then ice-cold ethanol is the preferred choice. You can chill it (in liquid nitrogen or at –80°C) to accelerate the precipitation without the risk of precipitating excess salt. Afterwards, you need to wash the pellet with 70% ethanol to remove any salt present.
Isopropanol is useful for large sample volumes e.g. the eluate you get after using a large volume plasmid kit. Because less isopropanol is needed for precipitation, you can often fit your sample and the solvent in one 15 ml tube.
However, because salts are generally less soluble in isopropanol than in ethanol, they tend to co-precipitate with DNA. To minimize the likelihood of salt precipitation, isopropanol precipitation is best at room temperature with short incubation times.
Once you recover the DNA or RNA pellet from the isopropanol, wash it with cold 70% ethanol to remove excess salt and to exchange the isopropanol for ethanol. It is ok to chill the isopropanol-precipitated sample if you are sure the sample doesn’t contain a lot of salt.
Because DNA is less soluble in isopropanol, isopropanol allows precipitation of larger species and lower concentrations of nucleic acids than ethanol, especially if you incubate at low temperatures for long periods of time.
If you do this, just remember to wash the pellet several times in 70% ethanol after pelleting, to reduce the amount of salt you carry over.
In Short – Ethanol or Isopropanol?
Use Ethanol If:
- You have the space to fit two volumes of ethanol to sample in your tube.
- The sample needs to be stored for a long period of time and will be chilled.
- You need to precipitate very small DNA fragments.
Use Isopropanol If:
- Your sample volume is large and you can only fit 1 volume of solvent into your tube.
- You need large molecular weight species.
- The DNA concentration in your sample is low.
- You are in a hurry and want to accelerate the precipitation of nucleic acids at room temperature.
Handy Tips
- Ethanol precipitation of DNA:
- Add 2 volumes of ethanol to the sample and freeze at –20°C for at least 1 hour or overnight for best results.
- Centrifuge the sample at full speed for 20 minutes to collect all material.
- Wash with 70% ethanol, then centrifuge for 10–15 minutes to pellet the DNA.
- Remember to mark the side of the tube where the pellet is expected to be and don’t let it out of your sight when decanting the ethanol!
- Isopropanol precipitation of DNA:
- Avoid cold temperatures because of the excess salt precipitation that can occur.
- To increase the yields precipitated, incubating the sample mixture at room temperature for longer periods rather than chilling the sample.
- When the DNA is pelleted, the pellet is sometimes more difficult to see compared to the ethanol pellet. It can be clear and glassy. Make sure, again, to note the side of the tube where the pellet should be. Look for it before decanting the isopropanol and 70% ethanol wash.
- After washing with ethanol, the pellet becomes visible and white. Make sure it doesn’t slip off the side of the tube wall before decanting the supernatant. Allow the tube to drain upside down for a few minutes, let it air-dry, or use a centrifugal evaporator (5 minutes is enough) and then resuspend in buffer.
- Finally, for dry DNA pellets, heating the sample in buffer at 50–60°C will help the DNA dissolve faster and won’t damage the DNA. Heating is also suitable for RNA, in a water bath at temperatures not exceeding 42°C. Over-dried DNA and RNA will take longer to dissolve so make sure not to evaporate for too long.
So now you know the difference between ethanol and isopropanol precipitation, and when to use each method. Good luck with your DNA precipitations!
References
- Green MR, Sambrook J. Precipitation of DNA with Isopropanol. Cold Spring Harb Protoc. 2017(8):pdb.prot093385.
Originally published on December 10, 2009. Updated and republished in August 2021.