Targeted protein degradation has rewritten the rulebook for drug discovery. Instead of blocking a protein’s function, degraders remove the protein itself.
Degrader compounds act as a bridge between a target protein and an E3 ligase. The resulting ternary complex (degrader + protein + ligase) enables the ligase to tag the protein with ubiquitin. The tagged protein is subsequently degraded in the proteasome (Figure 1). [1]
Unlike traditional inhibitors that must remain bound to a protein to keep it switched off, degraders only need to bind long enough to recruit the cell’s own degradation machinery. Once degradation occurs, the degrader is released and can engage with the next target.
Learning how to measure the kinetics of targeted protein degradation helps you understand why some degraders succeed where others fail, and guides you in designing better ones.
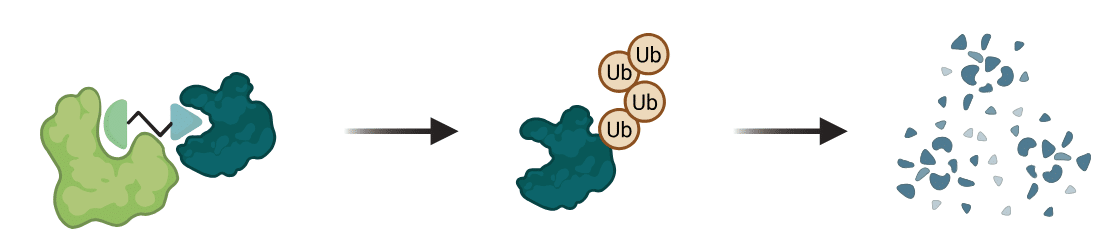
Figure 1. Targeted protein degradation exploiting the Ubiquitin Proteasome System. E3 ligase (light green) and a target protein (dark green) are brought together by a PROTAC degrader. The interaction results in the target protein being ubiquitinated and ultimately degraded.
The Power of Targeted Protein Degraders
The target protein will be cleared from the system once it is marked by the E3 ligase for destruction, but the effect persists because the degrader has been freed to engage with additional target proteins in the cell. This event-driven mechanism is what makes degraders so powerful.
The therapeutic potential is huge.
Because degraders eliminate proteins rather than block them, they can target proteins that are beyond the reach of traditional inhibitors, such as oncogenic transcription factors, mutant kinases without accessible binding pockets, and non-enzymatic scaffolding proteins. [2]
A single degrader molecule can induce the degradation of many target proteins, creating a catalytic effect that leads to strong potency at low doses.
Degraders also tend to produce longer-lasting effects, as the cell must synthesize new protein before function returns. These properties have spurred degraders into clinical development for cancer, inflammation, and neurodegenerative diseases. A clinical example is Vepdegestrant (ARV-471), an estrogen receptor degrader in Phase III clinical trials for the treatment of breast cancer. [3]
Why Measure the Kinetics of Targeted Protein Degradation?
Not all degraders behave the same. Two degraders might both achieve 90% target loss after 24 hours, yet behave differently over time. One could induce rapid, sustained depletion; another might act slowly or allow recovery within hours. These variations in how a protein’s degradation unfolds over time, collectively called protein degradation kinetics, can separate a promising degrader therapy from one that fails to translate.
A degrader’s success depends on more than just potency: optimal compounds show fast onset, deep and sustained target loss, and slow recovery once treatment ends. [2] Quantifying degradation parameters helps you understand not just if your degrader works, but how it works, giving you the insight needed to make it better.
Methods for Measuring the Kinetics of Targeted Protein Degradation
In the early days of protein degradation research, you would measure kinetics with multiple time point western blotting, time-consuming pulse–chase, or cycloheximide-chase assays. [4] These provided valuable information but lacked sensitivity, throughput, and temporal resolution.
Today, you can use a range of techniques to assess targeted protein degradation. However, the most precise insights into degradation dynamics can be achieved if you combine live-cell luminescent reporters with computational kinetic analysis.
Let’s walk through the main approaches.
1. Time-course experiments
A simple way you can map degradation kinetics is to track target loss over time.
In a cell-based time course, you expose cells to a fixed degrader concentration and collect samples at multiple intervals, typically spanning minutes to a full day. Measuring the remaining target protein by Western blot or an ELISA-based assay allows you to plot a degradation curve and calculate the degradation rate in cells.
These experiments are straightforward but typically lack the throughput and temporal resolution of newer live-cell reporter formats.
2. Using proteomics to quantify degrader activity
Mass spectrometry–based proteomics is a powerful way to measure changes in protein abundance across the proteome following degrader treatment.
Through time-resolved or multiplexed techniques, like tandem mass tags or data-independent acquisition, you can quantify thousands of proteins at multiple time points to reveal degrader selectivity, identify new substrates, and uncover off-target or compensatory pathway effects.
These data provide a global view of cellular responses and help you to connect target loss with downstream biological consequences.
While time-course proteomics can capture broad, population-level degradation dynamics, live-cell assays are generally more suitable for high-resolution kinetic analysis of individual targets.
3. Live-cell assays to measure and monitor protein degradation kinetics
You can monitor degradation processes in real time with modern live-cell assays. Two systems that have become widely used are Promega’s HiBiT and NanoBRET® PPI, both based on NanoLuc® luciferase technology.
HiBiT is an 11–amino acid peptide tag that is fused to your target protein. It has high affinity for its complementation partner, LgBiT. Upon HiBiT-LgBiT engagement, an active luciferase is formed that produces a bright luminescent signal in the presence of a substrate.
The emitted signal is proportional to the amount of tagged protein, with over 7 logs of linear dynamic range.
The small size and signal sensitivity make HiBiT ideal for CRISPR-based endogenous tagging to study target proteins under native expression conditions. DNA or mRNA LgBiT delivery enables intracellular expression of LgBiT, which supports live-cell assays that capture real-time protein degradation kinetics.
NanoBRET measures protein interactions rather than abundance. This assay detects bioluminescence resonance energy transfer (BRET) between a NanoLuc- or LgBiT complemented HiBiT-tagged protein (the donor) and a fluorescent acceptor linked to a ligand or degrader.
In degrader studies, changes in the signal reveal the efficiency and duration of the target–degrader–E3 complex formation inside cells, providing kinetic insight into ternary complex assembly and dissociation—two processes that heavily influence degradation rate.
Together, these systems provide speed, sensitivity, and quantitation to degrader research, allowing you to extract kinetic parameters that were once impossible to measure in living cells.
Kinetic Analysis and Parameter Extraction
Once you have obtained the protein degradation time-course data, the next step is to interpret the results. You can characterize degrader activity by fitting the time-dependent signal and extracting key kinetic parameters:
- Degradation rate constant (λ) — the speed of protein degradation
- Dmax — the maximal extent of degradation
- DC₅₀ — the degrader concentration that results in 50% of Dmax
- Recovery time (t₁/₂, rec) — the time for the protein to return to half of its baseline level after degrader removal
Plotting these parameters across concentrations and with different compounds provides a clearer picture of how each degrader performs. These data allow you to directly compare different degraders during structure-activity relationship and optimization studies.
Interpreting Degrader Kinetic Profiles
When you measure the kinetics of targeted protein degradation, you start to see recurring patterns that can reveal critical mechanistic details. Characteristic profiles that can guide optimization include:
- Classic kinetics: The optimal degrader profile demonstrates dose-dependent, targeted protein degradation that reaches Dmax within hours, followed by sustained suppression. This pattern suggests that efficient ternary complex formation, strong E3 ligase engagement, and effective ubiquitination occurred. [2]
- Hook effect: At higher degrader concentrations, activity paradoxically decreases, which occurs when excessive binary binding to either the E3 ligase or target prevents ternary complex formation—an important reminder that more isn’t always better. [2]
- Partial degradation: Protein abundance plateaus at an intermediate level, even with increased degrader concentrations. This kinetic profile often reflects structural or spatial limitations, like subcellular compartmentalization or access to only a subset of the protein pool. [2]
- Linear or slow kinetics: Degradation occurs gradually over time, sometimes linearly, though complete clearance of the target protein is rarely achieved by the degrader. Possible causes for slow kinetics include weak ternary complex cooperativity, low permeability, or inefficient ubiquitination. [2]
- Delayed kinetics: Degradation begins only after an initial lag phase. This delay can result from preferential off-target binding that triggers target degradation secondarily, or from poor permeability that delays the onset of degradation until the degrader accumulates to an effective intracellular concentration. [2]
- Rapid recovery kinetics: In some cases, target proteins are resynthesized rapidly, typically due to strong transcriptional feedback, a short degrader half-life, or rapid degrader efflux. Knowing how quickly recovery occurs helps predict how long the therapeutic effect will last and whether ongoing dosing is necessary. [2]
Each kinetic signature is shaped by multiple factors: degrader properties (cellular entry, linker length, ternary complex stability), cellular context (protein synthesis rate, localization, abundance of E3 ligase), and assay design.
Optimizing Degrader Design Using Kinetic Data
Once you have detailed kinetic data, you can use it to troubleshoot and refine degrader design. For example, adjusting linker length or modifying E3 ligase or target-binding ligands can shift a degrader’s slow or partial degradation activity towards rapid and sustained target loss. [5–7]
You can compare λ, Dmax, and recovery times across analogs to pinpoint where the process is limited.
Incorporating these kinetic readouts into the early stages of compound design allows you to fine-tune degrader potency, duration, and selectivity before advancing to preclinical testing.
How to Avoid Common Pitfalls When Measuring Targeted Protein Degradation Kinetics
Your well-designed assays can even yield misleading degradation curves if you don’t control for experimental artifacts. Compound instability, slow ternary complex formation, or transcriptional feedback are some of the factors that distort kinetic data.
To avoid these pitfalls, you can verify degrader stability over the assay timeframe, include non-functional analogs as controls, and measure protein resynthesis rates to confirm that the target’s return after degrader removal reflects genuine new protein synthesis rather than assay artifacts or signal interference.
It also helps to confirm that signal loss reflects targeted protein degradation rather than general cytotoxicity. Taking these steps ensures that kinetic differences reflect biology, not experimental noise.
From Measuring to Understanding the Kinetics of Targeted Protein Degradation
Degraders are powerful tools that control biology by rewriting protein lifetimes. To harness their full potential, though, you must understand not just whether they degrade, but how they degrade.
Combining live-cell assays, quantitative proteomics and kinetic analysis enables you to dissect each phase of the degradation process—from ternary complex formation to protein resynthesis—providing a complete picture of degrader behaviour.
By mastering how to measure the kinetics of targeted protein degradation, you can design degraders that act faster, last longer, and translate more effectively in the clinic.
References
- Sakamoto K. M. et al. (2001). Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F-box complex for ubiquitination and degradation. PNAS, 98(15), 8554–8559.
- Daniels D. L., Riching K. M., & Urh M. (2022). The importance of cellular degradation kinetics in understanding targeted protein degradation. Chemical Society Reviews, 51, 9429–9447.
- Arvinas Inc. (2025). Vepdegestrant (ARV-471) clinical trial update: Phase 3 results for advanced ER+ breast cancer. [Press release].
- Varshavsky A. (2008). Discovery of cellular regulation by protein degradation. Journal of Biological Chemistry, 283(50), 34469–34489.
- Churcher I. (2018). Protac-induced protein degradation in drug discovery: Breaking the rules or just making new ones? Journal of Medicinal Chemistry, 61(2), 444–452.
- Bondeson D. P. et al. (2015). Catalytic in vivo protein knockdown by small-molecule PROTACs. Nature Chemical Biology, 11, 611–617.
- Toure M. & Crews C. M. (2016). Small-molecule PROTACs: New approaches to protein degradation. Angewandte Chemie International Edition, 55(6), 1966–1973.









