You’ve just spent weeks nursing your organoids to perfection, and now you need to count them for your next experiment. But manual counting takes forever, your lab mate gets different numbers than you, and your old automated counter thinks half your beautiful spheroids are debris.
If that sounds familiar, then read on to find out how to count organoids accurately using machine learning-powered automation to make this step faster, more reproducible, and waste-free.
3D Cell Structures Are Amazing (but Frustratingly Hard to Count)
3D cell culture models mimic in vivo conditions far better than simple 2D cell culture. Examples include (Figure 1):
- Spheroids: simple 3D structures
- Tumorspheres: dense, tumor cell based
- Organoids: mimic in vivo most closely
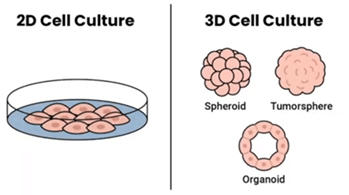
Figure 1. Comparison of 2D cell culture (left) with 3D cell culture examples (right).
Organoids are grown from stem or progenitor cells, which self-organize into dense 3D structures that look and act like miniature organs. They even have tissue-specific cell types. The cells have polarity, interact with one another and even with their environment, simulating what happens in native tissue.
Organoids are a big step towards representing in vivo conditions. That makes them incredibly useful for tissue engineering, regenerative medicine, drug testing for rare diseases, and cancer progression research. They are emerging as a more ethical and potentially more physiologically relevant complement or alternative to animal studies.
The problem is that manually counting them consistently is very difficult. The complexity of organoids creates problems, including:
- Irregular shapes and sizes: No two organoids look the same, so segmentation and size gating get messy fast.
- Variable density and brightness: Healthier or more compact structures show up differently.
- Overlapping and clustering: Individual boundaries blur together.
- Plain old subjectivity: Manual counts vary person to person, experiment to experiment.
The lack of a universal definition of what constitutes a “spheroid” or “organoid” adds another layer of inconsistency. One researcher might include small aggregates that another excludes. This subjectivity makes manual validation challenging and prevents meaningful comparison between labs.
When you miscount organoids, everything downstream (e.g. growth rates and treatment responses, the data your grant depends on) becomes unreliable. That’s why knowing how to count organoids accurately is really important.
Why Your Old Cell Counter Isn’t Cutting it
Most automated cell counters were designed for cell suspensions like mammalian cells or yeast. Their algorithms hunt for bright, circular objects and use simple thresholding to identify cells and calculate cell number and viability.
But 3D structures laugh in the face of those assumptions.
Organoids vary in density, shape, and optical properties. Figure 2 is an example of what your cell counter may be dealing with when you image 3D cell cultures. Older systems undercount clusters, mistake debris for tiny spheroids, or just give up entirely.
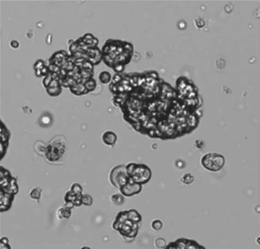
Figure 2. Brightfield image of lab-grown tumorspheres displaying irregular shapes and sizes.
Brightfield imaging of 3D cultures can also show uneven illumination and variable brightness depending on sample health and density, so automated algorithms mistake light patches for debris or miss faint outlines entirely.
So many researchers end up back at the microscope. It’s precise, but painfully slow and still subjective.
High-throughput organoid work needs a different kind of automation. One that recognizes complex morphologies and adapts to variable conditions without making you retrain it every Tuesday.
Machine Learning: Teaching Computers to See Organoids
One cell counter with software that has been trained on thousands of real organoid images, including healthy ones, weird ones, and all the dissociated cells and debris that clutter up a typical sample, is the CellDrop™ FLi counter from Denovix. Its patented DirectPipette™ technology replaces hemocytometers and plastic slides with a wipe-clean, height-adjustable chamber that you load your sample directly into.
To develop this cell counting software, Denovix had experts analyze thousands of images by hand, teaching the algorithm what an organoid looks like versus debris or cell fragments. After many rounds of retraining, the model learned to handle different 3D culture types and imaging conditions reliably.
This resulted in a pre-trained, ready-to-use app that classifies objects in seconds with no ongoing user training required (Figure 3). In under 30 seconds, you get standardized, reproducible counts.
The software distinguishes:
- Organoids (by outlining them in blue)
- Dissociated objects (by outlining in yellow)
- Debris (by outlining in red)
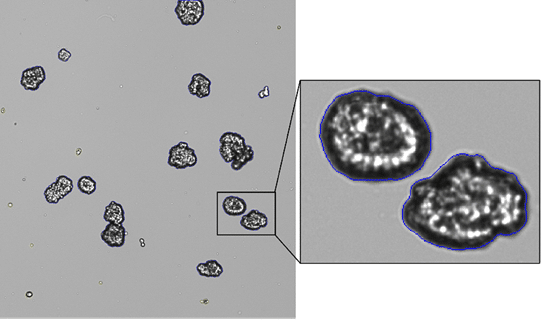
Figure 3. CellDrop FLi counter result image from the organoid app. Organoids are outlined in blue, and dissociated cells or fragments in yellow.
A Whole Organoid Counting Workflow in Under 30 Seconds
Counting organoids with the CellDrop FLi is very simple:
- Load your sample directly into the chamber with a pipette.
- Autofocus brings structures into view and remembers your settings for next time.
- Tap “Count.” Machine learning instantly classifies organoids, dissociated objects, and debris.
- Review data. View outlines, densities, mean diameters, and percentages in real time.
- Export and clean. Save as PNG, CSV, or PDF, then wipe the chamber and move on.
You can save protocols for specific organoid types, so every experiment starts from the same, consistent baseline. Figure 4 shows how you load a sample into the CellDrop FLi instrument.

Figure 4. CellDrop FLi user loading their sample directly into the instrument using a pipette.
Fine-tune it to Your Lab’s Standards
The CellDrop software lets you adjust analysis parameters, including object size and brightness thresholds—whatever matters to your definition of a “good” organoid. Once you’ve dialed it in, save those settings as a protocol for future use.
Real-time visualization tools let you plot size distributions or compare samples side by side, so you can track changes across passages or treatments without exporting to three different programs first.
Why Automating Organoid Counts Actually Matters
Automating organoid counts with machine learning and slide-free tech solves three problems at once:
Speed: You get reliable numbers in seconds, not after an hour hunched over a microscope.
Reproducibility: Counts stay consistent whether it’s you, your postdoc, or the new grad student doing the counting.
Sustainability: No disposable slides means less plastic waste and lower consumable costs.
For any lab running 3D culture systems, learning how to count organoids accurately is the difference between trustworthy data and questionable results.
Wrapping Up
If you’re working with spheroids, tumorspheres, or organoids, accurate quantification is the foundation of valid, reproducible science. Traditional counting methods can’t keep pace with the complexity of 3D models, but machine learning and sustainable workflows finally can.
If you incorporate a cell counter designed for 3D models, such as the CellDrop FLi, it automates the recognition work, handles the image analysis, and stays out of your way, so you can trust your organoid counts and move on quickly with the rest of your experiment.
To see the CellDrop FLi machine in action, watch our webinar with Application Scientist Grace Emin.