Consider a jigsaw puzzle. While most of the pieces have a different picture on their surface, all pieces fit together in an interlocking pattern.
As unlikely as it may seem, restriction enzymes from different organisms can produce interlocking pieces of DNA – so called compatible cohesive ends (CCE). These are pieces of DNA, which fit together and can be ligated, creating a hybrid molecule.
It’s all about isoschizomers
The recognition sequences of type II restriction enzymes used in molecular biology are palindromic, e.g. symmetrical. Further constraints on the restriction enzymes are the requirements to recognize certain bases in certain positions of a palindrome. The result is a finite number of the solutions for DNA recognition and cleavage puzzle – a lot of different species have unrelated enzymes, which recognize the same sequence (they are called isoschizomers).
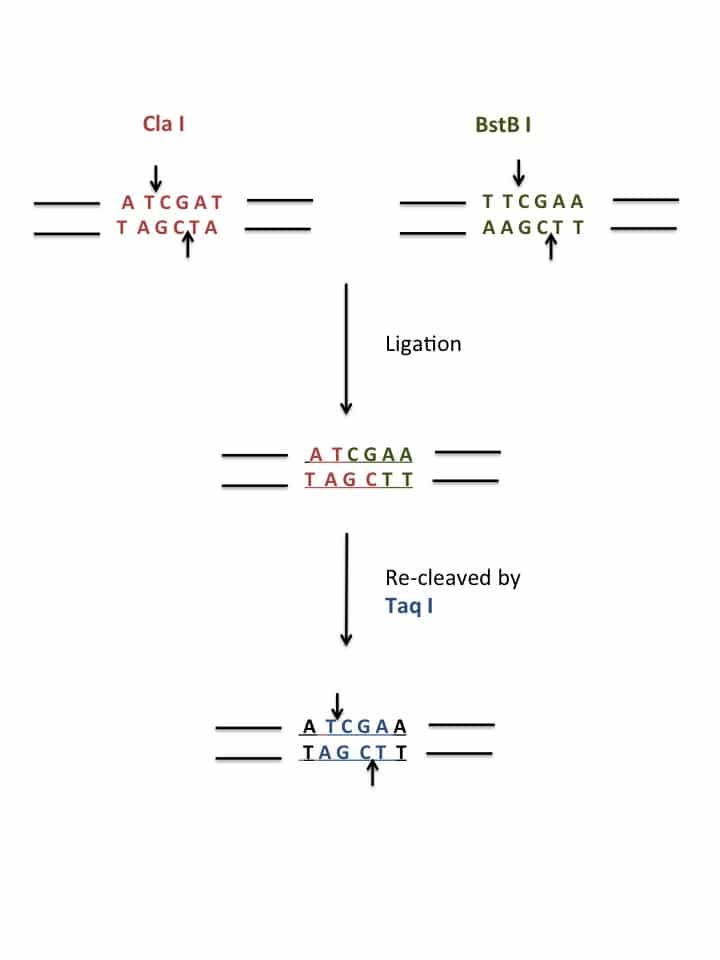
Of course, by definition, isoschizomers often produce compatible cohesive ends. However, unrelated enzymes with different recognition sequences can also produce compatible ends, which can be ligated together. Have a look at Figure 1 in which two different DNA molecules, cleaved by Cla I and BstB I, respectively, can be ligated together. In this case the resulting, hybrid molecule can be further re-cleaved by the third enzyme, Taq I. Conversely, ends cleaved by Taq I are compatible with Cla I and BstB I and several other enzymes.
Enjoying this article? Get hard-won lab wisdom like this delivered to your inbox 3x a week.

Join over 65,000 fellow researchers saving time, reducing stress, and seeing their experiments succeed. Unsubscribe anytime.
Next issue goes out tomorrow; don’t miss it.
Putting isoschizomers to work for you
You can see from Figure 1 that using CCE destroys both original recognition sequences.
So what’s the point in using CCE for cloning?
Well, sometimes it’s the only way to ligate, saving the necessity of a PCR.
More importantly, destruction of the restriction sites can be a useful diagnostic tool to distinguish between your vector self-ligation and the resulting construct containing the fragment of interest.
Identifying CCE
How would you know if your enzymes of choice produces CCE?
The simplest, low tech way is looking in the New England Biolabs catalog, which contains a lot of useful information in the last section. In the 2013-2014 edition the list of enzymes producing CCE, what they can be ligated to and how to re-cleave hybrid molecules is located on pages 312 – 314. Alternatively, NEB also provide this information on their website.
Let’s have fun with DNA jigsaw puzzle!
You made it to the end—nice work! If you’re the kind of scientist who likes figuring things out without wasting half a day on trial and error, you’ll love our newsletter. Get 3 quick reads a week, packed with hard-won lab wisdom. Join FREE here.