If you rely on bioluminescent assays or imaging—most often built around luciferase-based reporters—you already know the upside: bioluminescence gives you an incredibly clean background, wide dynamic range, and the ability to monitor low-level biology in real time.
But you also know the catch: bioluminescent signals are only as strong and stable as your assay design. Small variations in substrate handling, timing, instrument settings, or normalization can reduce sensitivity more than you might expect. And after spending all that time on your experiment, the last thing you want is noisy data you can’t trust.
In this guide, you’ll learn how to optimize bioluminescence assays for high-sensitivity detection, whether you’re measuring reporter activity in cell lysates, tracking live-cell kinetics, or performing in vivo imaging. Along the way, we’ll highlight practical considerations around enzymes, substrates, and instrumentation—and where specialized tools can help you achieve cleaner, more reproducible data.
Why Bioluminescence Enables High-sensitivity Detection
Here’s what makes bioluminescence so powerful: it’s generated inside your sample through a chemical reaction, usually between an enzyme (like luciferase) and a substrate (like luciferin). When luciferase catalyzes luciferin oxidation, you get light as a byproduct.
Unlike fluorescence, bioluminescence doesn’t require any external light stimulation. This results in less background fluorescence, light scattering, and photobleaching than will typically occur with fluorescence imaging. All this results in a cleaner signal.
In addition, most biological systems lack endogenous bioluminescent activity, meaning that cells, media, and tissues do not naturally emit light. As a result, nearly all detected signals in your assay originate from the reporter system itself. With a high signal-to-background ratio, you can detect subtle biological changes that other methods might miss and be confident that the signals have arisen from the reporter system you are studying.
Bioluminescence is also uniquely suited to live-cell imaging. Because this method doesn’t use excitation light, cells are spared the oxidative stress, membrane damage, and metabolic disruption caused by high-intensity illumination.
In practice, bioluminescent assays are far less phototoxic than fluorescence, making them gentler during long-term or high-frequency imaging. This allows you to monitor biological processes over extended periods of time without altering the behavior of the cells you’re studying.
These properties make bioluminescence a strong choice for applications where sensitivity and minimal perturbation are essential, such as:
- Low-abundance gene expression assays and reporter-based transcriptional readouts
- Live-cell assays tracking pathway dynamics and kinetics, including responses to drugs or stimuli
- Protein–protein interaction assays (think split luciferase complementation)
- High-dynamic-range reporter screening in multi-well formats
- In vivo bioluminescence imaging, where you need deep tissue penetration with minimal background
- Target engagement and protein degradation assays, such as PROTAC workflows
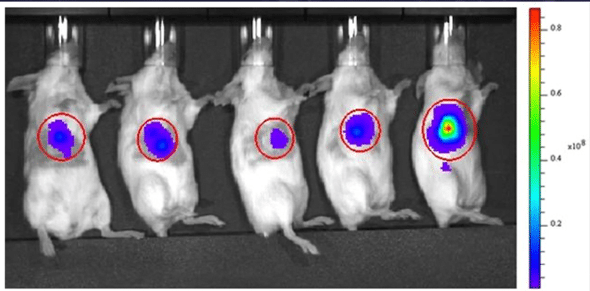
Figure 1. In vivo bioluminescence imaging of mice.
If you’d like a deeper dive into the fundamentals of luminescent chemistry and assay design, you can explore Promega’s Luminescence Resource Center for additional background and practical guidance.
How Luciferase Choice and Substrate Handling Affect Sensitivity
Not all luciferases work the same way.
- Firefly luciferase gives you a bright but short-lived flash of light, making precise timing essential.
- Stabilized glow-type luciferases produce flatter, longer-lasting signals that are a lot more forgiving in multi-well formats.
- ATP-independent luciferases, like NanoLuc® luciferase, produce an exceptionally bright and stable signal, making them ideal for low-level targets, protein fusion studies, and live-cell assays.
But here’s where it’s easy to trip up: substrate handling.
Luciferase substrates are finicky; they degrade with repeated freeze–thaw cycles and slowly oxidize over time. If you prepare fresh working solutions, properly store aliquots (protect them from light and follow the manufacturer’s recommended storage temperature, which varies by substrate), and mix thoroughly, you can significantly improve signal quality. Bubbles and uneven substrate delivery can also reduce sensitivity.
Temperature consistency matters as well. Luciferase activity is susceptible to temperature fluctuations, so let your plates and reagents equilibrate before reading. For ATP-dependent luciferases, keep your ATP and Mg²⁺ concentrations equal across wells.
Buffer composition plays a role, too. Detergents, pH differences, and inhibitors present in your lysis buffers can reduce enzyme activity. Standardizing your buffers helps maintain comparable performance between experiments.
Getting Your Timing and Reader Settings Right
Flash-type luciferases require precise timing because their signals peak and decay within seconds. You need an injector-equipped reader and consistent delay and integration settings. Every well needs to be read at the same time after substrate addition and measured for the same duration.
Glow-type luciferases are less dependent on millisecond timing than flash enzymes. However, they still require consistent incubation windows—you should read every well after the same amount of time has elapsed since you added the substrate. Variations in incubation windows can shift signal intensity and compromise reproducibility.
Now let’s talk instrumentation and equipment.
Use white microplates as they reflect emitted photons to the detector, significantly boosting your signal. Improved signal capture is especially important when working near the limit of detection.
Then you must ensure that your luminescence microplate reader is fit for purpose. Bioluminescent assays require a luminometer with high sensitivity and a broad dynamic range to detect very low levels of emitted light without saturating brighter signals.
Promega GloMax® microplate readers are sensitive, easy-to-use, and offer flexible configurations ranging from dedicated luminometers to advanced multi-mode systems.
You should also take time to optimize your reader’s gain, focal height, and integration time to achieve the maximum signal possible.
Normalization: The Difference Between Good Data and Great Data
You can’t skip normalization if you want to optimize bioluminescence assays for high-sensitivity detection. Here’s why normalization matters.
Dual-luciferase normalization is the most effective method for reporter assays. With this method, you use a second, constitutively expressed luciferase as an internal control for each well. Both luciferases experience the same transfection variability, cell number differences, and lysis efficiency.
When you divide the primary reporter signal by the control signal, you remove most of the technical noise and isolate the true biological response. Your data will become significantly more consistent across wells, plates, and experiments.
For live-cell assays, normalize your bioluminescent signal to something that accounts for cell number or well-to-well variation—total protein or DNA after lysis works, or a constitutive viability marker in intact cells. Plate-level corrections (like row/column adjustments or normalization to the plate median) can also reduce spatial artifacts and improve sensitivity.
Together, these approaches ensure that changes in luminescence reflect biology, not differences in cell density or the well’s position on the plate.
Taking High-sensitivity Detection In Vivo
In vivo bioluminescence imaging (BLI) is a non-invasive technique that lets you monitor cellular and molecular processes in real time in living organisms. Unlike other imaging modalities that may require contrast agents or radiation, BLI relies solely on the light produced by the bioluminescence reaction.
One major advantage of in vivo BLI is its non-toxicity. This means you can repeatedly image the same animal over time, tracking disease progression and gene expression patterns without harming your subject.
To optimize bioluminescence assays for high-sensitivity detection in vivo, you need three things:
- An animal model engineered to express a luciferase reporter gene (such as NanoLuc® or firefly luciferase)
- A luciferase substrate
- A sensitive imaging system
You typically administer the substrate by injection into the animal, where it reaches luciferase-expressing cells and, in the presence of oxygen and any additional enzyme-specific requirements, undergoes an enzymatic reaction that produces light. The specialized imaging system then captures this signal and converts it into a spatial map of reporter activity.
The signal intensity and distribution depend heavily on how efficiently the substrate is delivered, how much tissue the light must penetrate, and how consistently you handle and image your animals.
Substrate choice is crucial. For firefly luciferase models, standard D-luciferin works, but optimized formulations such as Promega’s VivoGlo™ Luciferin, In Vivo Grade give you more consistent imaging performance.
For NanoLuc®-based reporters, substrates such as Promega’s Nano-Glo® Fluorofurimazine In Vivo Substrate (FFz) provide better bioavailability for bright, stable signals.
These Promega substrates are spectrally compatible, so you can use them together for dual-luciferase imaging in whole animals, giving you more flexibility and more data.
Maintaining consistency in substrate dose, anesthesia conditions, animal temperature, and imaging timing helps stabilize signal across animals and time points.
For longitudinal studies, imaging at matched time points after substrate injection helps you control for pharmacokinetics and reveals true biological change rather than technical variation.
Normalization in animal studies typically involves controlling the region-of-interest size, matching imaging parameters, and comparing only animals processed under identical experimental conditions.
Bringing It All Together for High-sensitivity Bioluminescence Detection
Bioluminescence offers two defining advantages: essentially zero biological background and an exceptionally sensitive signal across a broad dynamic range.
When you optimize bioluminescence assays for high-sensitivity detection—from luciferase selection and substrate handling to timing, reader configuration, and normalization—you can reliably detect subtle biological events that would otherwise remain hidden.
Combined with reagents designed specifically for live-cell and in vivo applications, these optimizations help you generate cleaner, brighter, and more reproducible bioluminescent data across both cell-based and whole-animal systems.