The completion of the Human Genome Project in 2003 ushered in a new era of rapid, affordable, and accurate genome analysis—called Next Generation Sequencing (NGS). NGS builds upon “first generation sequencing” technologies to yield accurate and cost-effective sequencing results.
Fred Sanger sequenced the first whole DNA genome, the virus phage ?X174, in 1977. In that same year, Sanger developed the future backbone of the genome era: DNA sequencing technology. His technique used the chain termination method. This is now seen as the “first generation technology” of genome sequencing.
Since Sanger sequencing (or the chain termination method) is the first generation of sequencing technology, understanding it is greatly important. The Human Genome Project began with Sanger sequencing technology.
The Sanger Sequencing Method
The Sanger sequencing method relies on dideoxynucleotides (ddNTPs),a type of deoxynucleoside triphosphates (dNTPs), that lack a 3′ hydroxyl group and have a hydrogen atom instead . When these bases bind to the growing DNA sequence, they terminate replication as they cannot bind other bases. To perform Sanger Sequencing, you add your primers to a solution containing the genetic information to be sequenced, then divide up the solution into four PCR reactions. Each reaction contains a with dNTP mix with one of the four nucleotides substituted with a ddNTP (A, T, G, and C ddNTP groups). At the end of the PCR, each of your four reactions will yield PCR products of various lengths because replication is randomly terminated. By running the samples on a gel with 4 lanes, you can piece together the sequence as each sequence has been replicated from the same original material. Here is an example where the ddNTPs are in bold and the dNTPs are not:
Enjoying this article? Get hard-won lab wisdom like this delivered to your inbox 3x a week.

Join over 65,000 fellow researchers saving time, reducing stress, and seeing their experiments succeed. Unsubscribe anytime.
Next issue goes out tomorrow; don’t miss it.
Your sequence is ATGCTCAG.
Your four reactions give you:
Reaction with ddATP: A, ATGCTCA, ATGCTCAG
Reaction with ddGTP: ATG, ATGCTCAG
Reaction with ddCTP: ATGC, ATGCTC, ATGCTCAG
Reaction with ddTTP: AT, ATGCT, ATGCTCAG
All the reactions once run a gel would look something like this (Image by Olwen Reina):
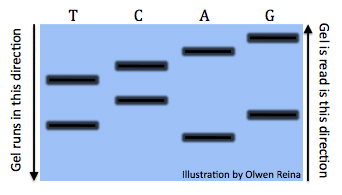
Each band denotes the different lengths code. For example, the band is the right under the “A” symbolizes the sequence: “ATGCTCA”
Still confused?
Let’s imagine a party game. The game is a guessing game. Here is how it is played:
You are thinking of a number and the group has to guess it. The tricky part is that the number is 200-digits in length. You are reading the digits of the number in your head without making a sound. Every so often a person interrupts you, and you tell them the single digit you were just thinking and where it is in the sequence of 200. Each time you are interrupted, you have to start again. You leave after a few hours and the group has to figure out the 200-digit number. They have to piece together the information you gave them, for example the 25th number was 5, the 40th number was 0, and so on. Using the information from their interruptions, they can repeat the number they gave you.
While this sounds like the lamest game in the world, it works very well for sequencing!
Unfortunately, it is slow, expensive, and (previously) relies on radioactive materials. This pushed scientists to develop new and better forms of genome sequencing.
Next Generation Sequencing Technologies
The biggest advances in genome sequencing have been increasing speed and accuracy, resulting in reduction in manpower and cost. The speed is thanks to parallel analysis and high throughput technology. It’s the difference between getting a 4 year-old to read Moby Dick and giving a paragraph each to a bunch of Drama majors and asking them to read at once.
Sequencing machines have improved wildly since Sanger developed his method. In 1987, Applied Biosystems became the first to introduce an automatic sequencing machine, called the AB370. The machine relied on a method called capillary electrophoresis that used Sanger’s chain terminating method without needing a gel.
Fluorescently labelled chain-terminating ddNTPs are added to the PCR reaction mix. By the Sanger sequencing method, PCR products of various lengths are created, and then separated according to their size. Size is measured by the PCR product’s overall negative charge. The more negative the charge, the longer the fragment. Take the above example of ddNTP incorporation and imagine that the bold letters are ddNTPs with a fluorochrome attached. As the fragments are pulled toward the positive electrode of a capillary (see image below), they pass a laser beam that triggers a flash of light from the fluorochrome attached to the ddNTP that is characteristic of the base type (for example, green for A, yellow for T, blue for G, red for C). In this way, the genome is carefully read.
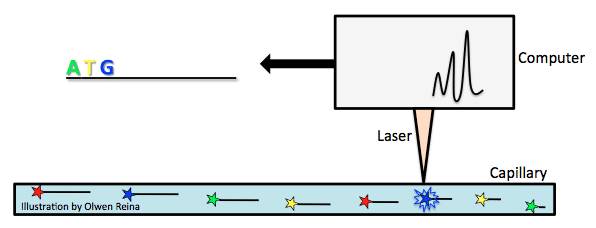
The biggest difference here was speed and cost. AB370 could detect 96 bases at one time, 500K bases per day, and had a read length of 600 bases by using a parallel analysis and high throughput setup. This form of sequencing became the main tool for the completion of the human genome project.
How Does NGS Compare to First Generation Sequencing?
NGS is characterized by improved accuracy and speed, but also reduced manpower and cost. There has never been a time where it has been as cheap, convenient, or straightforward to sequence a genome. Arguably, the biggest improvement has been the development of parallel analysis, which increased the sequencing speed.
The only thing slowing us down now is the interpretation of results!
You made it to the end—nice work! If you’re the kind of scientist who likes figuring things out without wasting half a day on trial and error, you’ll love our newsletter. Get 3 quick reads a week, packed with hard-won lab wisdom. Join FREE here.







