How To Identify Conserved Elements In Genes
Conserved elements are stretches of DNA sequence that are under purifying selection. That means mutations leading to a change of function in this part of the DNA are detrimental to the organism and will not become fixed in the genome, but rather discarded by natural selection. The level of conservation between species gives an idea of the relative “importance” of a stretch of DNA. For example, coding regions of genes with basic functions such as the cytoskeleton component B-actin are conserved between species as far apart as human and hydra, while genes involved in neural development have evolved a variety of different functions that is reflected in the divergence of their sequence. To identify conserved elements in your gene of interest, you will need to align the same sequence from several species and look for areas that they have in common. In this article, we’ll discuss how to choose the sequences you include in your alignment and how to spot the conserved elements.
Relationships between genes
It is important to be aware of the types of relationships genes can have both within and between species. Within species, gene duplications and subsequent diversification lead to gene families that often share a core conserved element, such as a homeodomain or basic Helix-loop-Helix domain that defines this group of genes. Recent copies of the same gene that occurred by whole genome duplication, such as at the rise of vertebrates or in the teleost fish lineage, result in paralogs. Paralogs are two closely related duplicates that occur in the same genome and have a letter or number added to their name that distinguishes them, such as pax6a, pax6b. Between species, the direct equivalent of one gene is called a homolog. Homologs usually carry the same name including a species identifier, such as xath5 (Xenopus ath5) and math5 (mouse ath5). To complicate things even further, a paralog in one species is called the ortholog of the paralog in another species (see figure 1). It is helpful to be aware of how your gene sequences are related even before you try to align them, because the alignment will be far easier to understand.
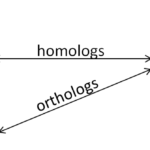
Figure 1: Nomenclature of relationships between genes and their duplicates within and between species.
Relationships between species
Choosing the right species within which to compare sequences depends largely on the question you want to answer. When making a phylogeny of a gene, it is often of interest to find out which conserved or functional domains of the gene were present in the ancestral species, and most of the time you will know what species or group of species you want to investigate. For example, if you are interested in the conservation of a gene within mammals, the outgroup should be a lower vertebrate with a comparable copy number of the gene and an uncomplicated genome. Watch out for whole genome duplications (teleost fish) or polyploidy (Xenopus laevis) which will potentially complicate your alignment. Additionally, you should add a well annotated model species as a reference (mouse, human, fruitfly for invertebrates).
Visualizing your alignment
Several good freeware alignment editors exist for most operating system. Jalview is a very easy to use viewer/editor that will color your alignment according to percentage similarity and additionally shows you conservation at each position of the alignment and a consensus sequence. Seaview is a similar editor that gives you many additional features like phylogenetic analysis of many kinds and can also perform the initial alignment for you. A helpful feature is the program Gblocks (available online or integrated in Seaview) which removes long stretches of gaps in your alignment, thereby streamlining it to more conserved regions. This should, however, be used with care, since parts of your sequences will be deleted.
Spotting conserved regions
Using a color scheme that tints every nucleotide in a different color makes it easy to spot highly conserved regions. Differences between species will often occur only in the so-called wobble bases, the third base of each three-nucleotide codon, or in single nucleotide polymorphisms (SNPs). Generally, the further apart the species are in which you find conservation, the more important the function of the specific part of the protein will be. After you have located conserved areas in your gene, you can use them to look for functionally important mutations between species or as a template for degenerate primers.
Useful links
Jalview: https://www.jalview.org/
Seaview: https://pbil.univ-lyon1.fr/software/seaview.html
Gblocks: https://molevol.cmima.csic.es/castresana/Gblocks.html
2 Comments
Leave a Comment
You must be logged in to post a comment.
Thanks for sharing!Learned.
There is a typo in your figure: two “Species 1”.