Mysterious miRNAs: An Introduction to MicroRNAs
mRNA, miRNA, siRNA, tRNA, rRNA! Just what do all these RNAs do? Most biology graduates will have heard a good deal about mRNAs, tRNAs and rRNAs since these are vital players in protein synthesis. For siRNAs there has been a lot of focus within drug discovery and biomedical research over the last decade, but there has been somewhat less focus on microRNAs (at least from a teaching perspective).
This short series of articles aims to take you on a journey through the miRNA world. In this first article, we will introduce microRNAs in general terms, looking at how they are generated and how they work. In the follow up articles, we will go into more detail, looking at how research can identify new miRNAS, and their targets.
A microRNA (miRNA) is a small non-coding RNA molecule, ranging typically from 21–25 nucleotides in length. miRNAs are widespread throughout the plant and animal kingdoms, and are found in some viruses. miRNAs are remarkably conserved in both plants and animals, and are thought to be a vital and evolutionarily ancient component of genetic regulation.
Discovery of miRNAs
miRNAs were first described in the journal Cell in 1993 by Lee et al. at Harvard University following their work in the model nematode, C. elegans.1 While studying postembryonic development in C. elegans, they made the fascinating discovery that lin-4, a gene previously known to negatively regulate the lin-14-encoded protein, did not actually encode a protein. Instead, they found strong evidence that lin-4 encoded 2 short transcripts of approximately 22 and 61 nucleotides in length and that these transcripts contained sequences complementary to the 3-UTR (untranslated region) of lin-14 mRNA.1 These exciting observations suggested that lin-4 regulated lin-14 translation by an antisense RNA mechanism and for many years this type of antisense regulation was believed to be a phenomenon restricted to nematodes.
MiRNAs Aren’t Just for Nematodes
Fast forward 6 or 7 years, and the second miRNA is found. let-7 was found to negatively regulate lin-41 mRNA to promote a later developmental transition in C. elegans. These findings were published in Nature by Reinhart et al.2 from Harvard Medical School, and soon afterwards the let-7 gene was found in a range of diverse organisms, prompting researchers to investigate the possible role for this new class of RNA molecules in developmental timing in humans. In the years that followed, this class of novel RNAs had been extended to include many members, but expression profiling revealed that they were probably not involved in developmental regulation in humans, but were likely to be involved in other regulatory pathways. The term miRNA was coined by Gary Ruvkun in 20013, and the field of miRNA biology was officially born.
Many miRNAs Means Many Targets
Since then, the human genome has been predicted to encode over 1000 miRNAs and many of these have been identified through a combination of random cloning and sequencing or computational prediction. Many of the miRNAs studied to date are abundant in a range of mammalian cell types, and collectively, they appear to target about 60% of known animal genes! The majority of known miRNAs are thought be to be involved in very distinct processes such as immunity, muscle biology and development, and aberrant miRNA expression has been implicated in a number of diseases including cancers, heart disease, and metabolic disorders. The miRBase, hosted by the Sanger Institute, is a useful resource with up-to-date information on miRNA nomenclature, nucleotide sequence, annotation and target/function predictions.
How Are MiRNAs Generated?
Plant and animal miRNAs are encoded by eukaryotic nuclear DNA, while viral miRNAs are encoded by the viral DNA of DNA viruses. The next paragraphs will focus mainly on animal miRNAs.
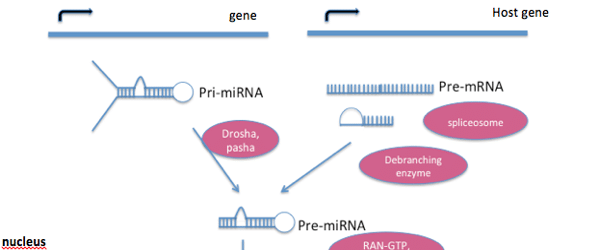
The genes encoding miRNAs tend to be much longer than the processed mature miRNA molecule. Many miRNAs are known to reside in the introns of their pre-mRNA host genes, sharing their regulatory elements and primary transcript, and both genes generally have a similar expression profile. For the remaining miRNA genes that are transcribed from their own promoters, only a few primary transcripts have been fully identified. A typical animal miRNA is generated as follows (See Figure 1 for illustration):
- MicroRNAs are transcribed by RNA polymerase II as large RNA precursor molecules called pri-miRNAs. Pri-RNAs comprise a 5′ cap and a 3’ poly-A tail.
- Pri-miRNAs are processed in the nucleus by a microprocessor complex, made up of the RNase III enzyme Drosha, and the dsRNA-binding protein, Pasha. The processed pre-miRNAs are approximately 70-nucleotides long and are folded into imperfect stem-loop structures.
- Pre-miRNAs are subsequently exported into the cytoplasm by the exportin 5 (Exp5) and Ran-GTP complex. Ran (ras-related nuclear protein) is a small GTP-binding protein that is essential for RNA and protein translocation through the nuclear pore complex. The Ran GTPase binds Exp5 and forms a nuclear heterotrimer with pre-miRNAs.
- In the cytoplasm, the pre-miRNAs undergo further processing by the RNAse III enzyme Dicer generating the final miRNA of approximately 21–25 nucleotides in length. Although either strand of the miRNA duplex can potentially have miRNA activity, only one strand is usually incorporated into the RNA-induced silencing complex (RISC). RISC is sometimes referred to as miRNP (miRNA ribonucleoprotein complex) and it is here that the miRNA and its mRNA target interact. The Dicer protein initiates the formation of RISC. RISC also is responsible for the gene silencing observed due to RNA interference.
Figure 1. Processing of a typical animal miRNA
The miRNA is transcribed from its own gene or a host gene (generally from within an intron) as a long pri-miRNA transcript. This is processed to pre-miRNA, which is then transported to the cytoplasm by Exportin-5, where it undergoes further modification by Dicer forming the finished miRNA molecule, ready for gene silencing. Image redrawn from Narayanese.
How Do MiRNAs Function?
miRNAs function by base-pairing complementary sequences within mRNA molecules, and this results in sequence-specific silencing of an mRNA molecule through one of the following processes:
1) cleavage of the mRNA strand into two pieces.
2) destabilization of the mRNA through shortening of its poly(A) tail.
3) less efficient translation of the mRNA into proteins by ribosomes.
In the next article we will cover how you can go about identifying miRNAs in your own research.
References:
- Lee RC, et al. (1993) ? The C. elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to lin-14. Cell. 75:843–854.
- Reinhart BJ, et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 403:901–6.
- Ruvkun, G. (2001). Molecular Biology: Glimpses of a Tiny RNA World. Science. 294:797–799.
2 Comments
Leave a Reply Cancel Reply
You must be logged in to post a comment.

[…] Our understanding of the biogenesis of miRNAs has improved over the years. Nowadays, it is well accepted that plant miRNAs are approximately 21 nucleotides long and are processed from stem-loop regions of long primary transcripts by a DICER-like enzyme prior to loading into silencing complexes. Once loaded into these complexes, they generally direct cleavage of complementary mRNAs, similarly to animal miRNAs. […]
[…] my first article on this topic we delved into what miRNAs are, how they are generated, and their function. Now, we […]