Immunophenotyping: Identifying Who’s Who in the Cellular World
Figuring out what’s what
When studying cells and cell subsets (and cell sub-subsets, and so on!!) we need ways to identify and classify every single cell. This will allow us to individually analyse each population and, for example, help to discover their role in health and disease. A principal way we do this is by looking at the proteins that the cells are expressing; usually on the cell surface but also inside, and the fancy term for all this is immunophenotyping.
How immunophenotyping works
The most straightforward way to profile the cells is to use antibodies that bind to the proteins of interest. Since specific proteins can be expressed on more than one cell type, we usually need to analyse quite a few at once. So what technology will suit this type of analysis? Well flow cytometry is a technique that allows analysis of multiple parameters on single cells, at rates of thousands of cells per second. So a perfect match or what?
With tons of reagents to choose from, you have almost your pick of monoclonal antibodies conjugated to hosts of different fluorescent molecules – which makes it all the more easy for you to look into these subsets. To make things easier in the long run you should start by spending a little time deciding on what proteins you’re interested in, and what fluorescence molecules will work best together in your mix, along with the instrumentation you have available. Remember to make sure your cytometer is equipped to analyse your selected fluorochromes; e.g. no red laser equals no APC (Allophycocyanin) detected!
It’s what’s on the outside that counts? CD molecules on the cell surface
On the protein side, one of the main things we talk about are CD (cluster of differentiation) markers. CD molecules are cell-surface proteins; mainly involved in cell communication, metabolism, or adhesion. Back in the day someone had the good sense to organise workshops every few years where folk would discuss such proteins, their function, and what cells you’d find them on, and then went about categorising them all. Currently there are over 330 CD markers listed (November 2014), and we usually classify cells by these CD markers, frequently along with the cells’ forward and side scatter characteristics. Some cell types can be characterised by the expression of just one or two (e.g. CD19+ B Cells, CD3+CD4+ helper T Cells), whilst others are a bit trickier, requiring analysis of many markers at once, and then examining their relative expression levels (e.g. human neutrophils are characterised by CD15+, CD16high, CD14lowCD52low).

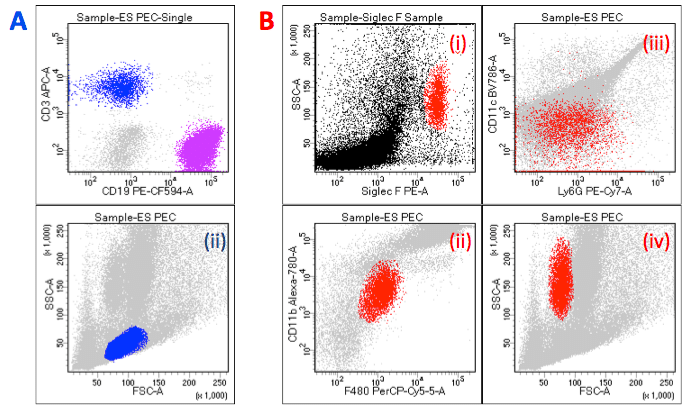
Example of immunophenotyping plots: A: Simply classification of T Cells [in blue] (CD3+) and B Cells [in purple] (CD19+) (i); with lymphocytes [in blue and purple] distinguished by Forward and Side Scatter (ii).
B: Classification of eosinophils [in red] as SiglecF+ (i) F4/80intCD11bint (ii) Ly6G-/lowCD11c– (iii) FSCloSSCint/hiSiglecF+ (iv).
(Note SiglecF antibody should not be used in cell sorting as antibody binding triggers cell death!) All plots represent sample from murine Peritoneal Exudate Cells.
Beneath the surface
As well as the cell surface, we can also take a look inside the cell to see what’s going on in there, and can use intracellular staining to tag those proteins. However, for this we have to fix all the proteins in place, and then permeabilise the cell membrane in order to get the antibodies in. In doing so we’re going to kill all the cells along the way – no big deal if we just want to take a look, but not much good if we want to do something with the cells afterwards, like purify and re-culture.
Beware!
Some caveats (aren’t there always?) to consider and try to control for:
- Activated cells can increase or decrease the levels of some proteins on or inside the cell (e.g. CD25 is expressed on regulatory T cells, but also on CD4+ T cells when activated).
- Specific antibodies may kill cells (e.g. Siglec F is specific to eosinophils but induces apoptosis upon binding) .
- Freeze-thawing or certain gradients can sometimes change the protein expression (e.g. CD62L can appear significantly higher following Ficoll-Hypaque density separation).
- Enzyme treatments to dissociate cells can lead to the shearing off of some of the proteins.
- Antibodies might not stick to where they’re supposed to, might tag onto the wrong cells, or get stuck in dead cells.
Immunophenotyping has greatly empowered life science research, and allows us to figure out what’s what, in particular when we’re looking at a mixed bag of cells; so then we can go about learning what they’re all up to. For example, looking at some cancers, and their severity and prognosis, we can use immunophenotyping to tell us what the unwelcome cell type is, at what stage it’s at, how aggressive it is, and if it’s getting better or worse. In various conditions, we may see more of one or another cell type, or see protein up/down regulation. We can use additional markers (e.g. intracellular cytokine staining) to look into the function of the subsets, and we can even purify the sub-populations out (by magnetic or cytometric cell sorting) and see how they behave on their own.