Navigating the sea of human genetics, Part II

Avast, me hearties, I’m back with Part II of our exploration of the HuGE Navigator. To get up to speed, be sure to check out Part I.
In Part II I’m going to go over the HuGEtools, which are used to mine the human genome epidemiology literature. The tools are:
- HuGE Literature Finder
- HuGE Investigator Browser
- HuGE Watch
- Variant Name Mapper
- HuGE Risk Translator
- Gene Prospector
Let’s go over them one by one.
HuGE Literature Finder contains data collected since 2001 (so keep in mind this limitation). It is a search engine for identifying information in the published literature on human genome epidemiology.
The search query can include genes, disease, outcome, environmental factors, author, etc. Results can be filtered by these categories. It is also possible to see all articles in the database for a particular topic, such as genotype prevalence, pharmacogenomics, or clinical trial.
Our Part I search example of “seasonal affective disorder”, aka SAD, brings up 15 articles that can be filtered by 7 genes.
HuGE Investigator Browser helps you search for investigators on human genome epidemiology topics by first/last or all authors. This info is obtained using a behind-the-scenes tool that automatically parses PubMed affiliation data.
Our SAD search results in 17 first/last authors and a total of 73 investigators. Results can be filtered by country or institute.
HuGE Watch helps you track the evolution of published HuGE research.You have a choice to view the trends in the HuGE published literature, HuGE investigators, genes studied, or diseases studied.
For example, if you search “Trend/Pattern” for “Diseases Studied” you’ll initially get a graph and chart of the number of diseases studied per year since 1997. You can refine these results by limiting the temporal trend to a category or study type such as “Gene-gene Interaction” or “HuGE Review”.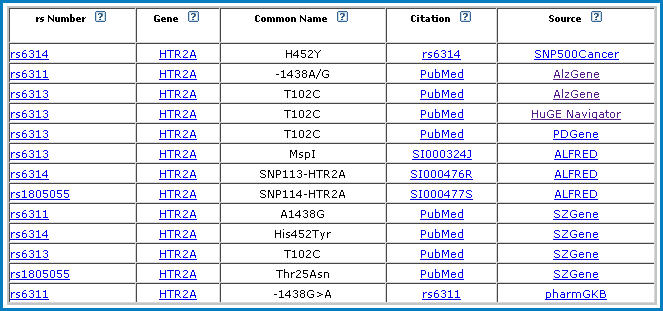
Variant Name Mapper lets you map common names of gene variants to theirappropriate rs numbers using information from SNP500Cancer, SNPedia, pharmGKB, ALFRED, AlzGene, PDGene, SZgene, HuGE Navigator, LSDBs, and user submissions.
As an example, one of our SAD genes, HTR2A, mapped to 4 different rs numbers, with 9 different common names, from 7 different sources.
HuGE Risk Translator calculates the predictive value of genetic markers for disease risk. To do so, users must enter the frequency of risk variant, the population disease risk, and the odds ratio between the gene and disease. This information is necessary in order to yield a useful predictive result.
Gene Prospector is “a gateway for evaluating genes in relation to disease and risk factors.” This extremely cool tool allows you to enter a disease or risk factor and then supplies you with a table of genes associated w/your query that are ranked based on strength of evidence from the literature. This evidence is culled from the HuGE Literature Finder and NCBI Entrez Gene. And to show you that it’s all above board, you’re even given the scoring formula.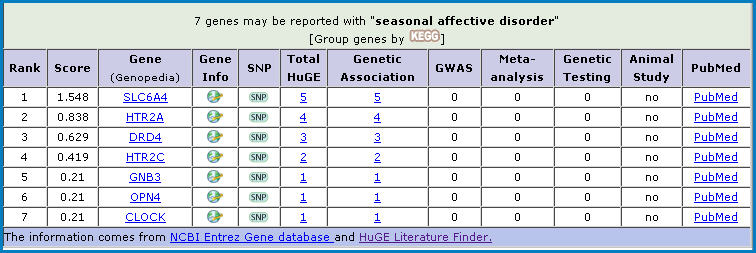
The Gene Prospector results table provides access to the Genopedia entry for each gene in the list, general info including links to other resources, SNP info, and associated literature from HuGE, PubMed, GWAS, and more. It is a great place to locate a lot of info about your disease/gene of interest very quickly.
To find out more about Gene Prospector, read this article by Wei Yu et al in BMC Bioinformatics.
So now you know the ropes, climb aboard the HuGE Navigator and get ready to explore brave new worlds of genetic associations and human genome epidemiology.
May you find bountiful treasures.
You can discuss your voyages on the HuGE Navigator, or ask any questions in our forum. Use the form below or go to the forum itself to start a new discussion.